6KSF
 
 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | 著者 | Li, H, Zhang, M. | | 登録日 | 2019-08-23 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
1FJA
 
 | |
6FB5
 
 | |
5UUG
 
 | |
5CL7
 
 | |
5CL4
 
 | |
5CL5
 
 | |
5CL6
 
 | |
5CDP
 
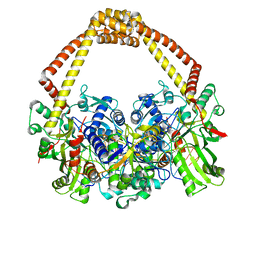 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5VVL
 
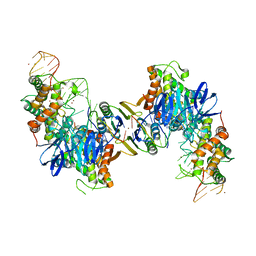 | | Cas1-Cas2 bound to full-site mimic with Ni | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (11-MER), ... | | 著者 | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | 登録日 | 2017-05-19 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
146D
 
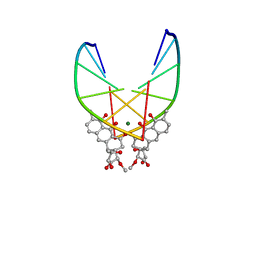 | | SOLUTION STRUCTURE OF THE MITHRAMYCIN DIMER-DNA COMPLEX | | 分子名称: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*CP*GP*CP*GP*A)-3'), ... | | 著者 | Sastry, M, Patel, D.J. | | 登録日 | 1993-11-09 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the mithramycin dimer-DNA complex.
Biochemistry, 32, 1993
|
|
6GDN
 
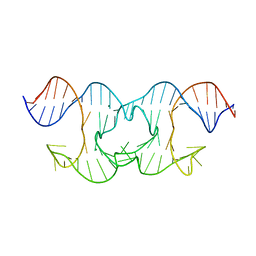 | | Holliday Junctions formed from Telomeric DNA | | 分子名称: | MAGNESIUM ION, Telomere DNA (42-MER) | | 著者 | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | 登録日 | 2018-04-24 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
5VVJ
 
 | | Cas1-Cas2 bound to half-site intermediate | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | 著者 | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | 登録日 | 2017-05-19 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.89 Å) | | 主引用文献 | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5UUH
 
 | |
3NT7
 
 | |
1CRX
 
 | | CRE RECOMBINASE/DNA COMPLEX REACTION INTERMEDIATE I | | 分子名称: | CRE RECOMBINASE, DNA (5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*G)-3'), ... | | 著者 | Guo, F, Gopaul, D.N, Van Duyne, G.D. | | 登録日 | 1997-07-02 | | 公開日 | 1998-10-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse.
Nature, 389, 1997
|
|
1MUG
 
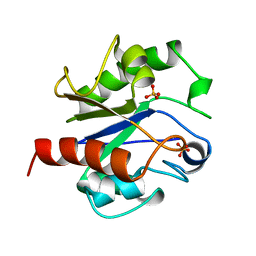 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | 分子名称: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | 著者 | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | 登録日 | 1998-07-10 | | 公開日 | 1998-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
7UJZ
 
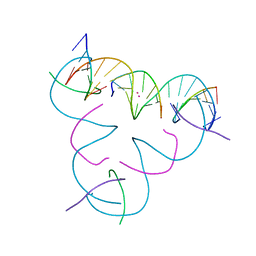 | | [T:Cd2+/Hg2+:T--pH 11] Metal-mediated DNA base pair in tensegrity triangle | | 分子名称: | CADMIUM ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-03-31 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.94 Å) | | 主引用文献 | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
159D
 
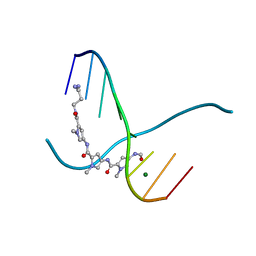 | | SIDE BY SIDE BINDING OF TWO DISTAMYCIN A DRUGS IN THE MINOR GROOVE OF AN ALTERNATING B-DNA DUPLEX | | 分子名称: | DISTAMYCIN A, DNA (5'-D(*IP*CP*IP*CP*IP*CP*IP*C)-3'), MAGNESIUM ION | | 著者 | Chen, X, Ramakrishnan, B, Rao, S.T, Sundaralingam, M. | | 登録日 | 1994-02-10 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Nat.Struct.Biol., 1, 1994
|
|
1SSP
 
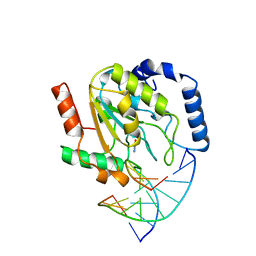 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | 分子名称: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | 著者 | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | 登録日 | 1999-04-28 | | 公開日 | 1999-05-06 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
5V9X
 
 | | Structure of Mycobacterium smegmatis helicase Lhr bound to ssDNA and AMP-PNP | | 分子名称: | ATP-dependent DNA helicase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Ordonez, H, Jacewicz, A, Ferrao, R, Shuman, S. | | 登録日 | 2017-03-23 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Nucleic Acids Res., 46, 2018
|
|
4LIL
 
 | | Crystal structure of the catalytic subunit of human primase bound to UTP and Mn | | 分子名称: | DNA primase small subunit, MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE, ... | | 著者 | Vaithiyalingam, S, Eichman, B.F, Chazin, W.J. | | 登録日 | 2013-07-02 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insights into Eukaryotic Primer Synthesis from Structures of the p48 Subunit of Human DNA Primase.
J.Mol.Biol., 426, 2014
|
|
6IFM
 
 | |
199D
 
 | | Solution structure of the monoalkylated mitomycin c-DNA complex | | 分子名称: | CARBAMIC ACID 2,6-DIAMINO-5-METHYL-4,7-DIOXO-2,3,4,7-TETRAHYDRO-1H-3A-AZA-CYCLOPENTA[A]INDEN-8-YLMETHYL ESTER, DNA (5'-D(*(DI)P*CP*AP*CP*GP*TP*CP*(DI)P*T)-3'), DNA (5'-D(*AP*CP*GP*AP*CP*GP*TP*GP*C)-3') | | 著者 | Sastry, M, Fiala, R, Lipman, R, Tomasz, M, Patel, D.J. | | 登録日 | 1994-12-01 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the monoalkylated mitomycin C-DNA complex.
J.Mol.Biol., 247, 1995
|
|
7V59
 
 | | Cryo-EM structure of spyCas9-sgRNA-DNA dimer | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (49-MER), RNA (115-MER) | | 著者 | Liu, J, Deng, P. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (5.26 Å) | | 主引用文献 | Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search.
Chem Sci, 12, 2021
|
|
