2NCN
 
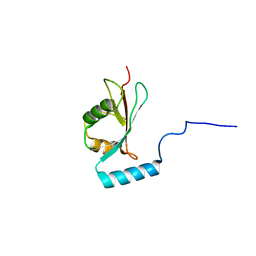 | |
2NCO
 
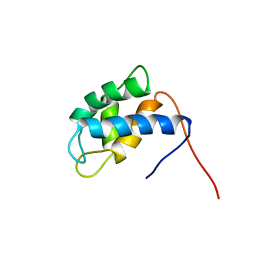 | |
2NCP
 
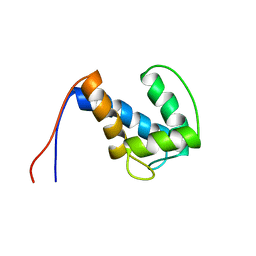 | |
2NCQ
 
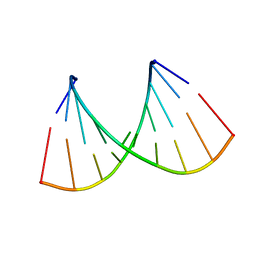 | |
2NCR
 
 | |
2NCS
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | 分子名称: | Envelope glycoprotein gp41 | | 著者 | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | 登録日 | 2016-04-14 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2NCT
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | 分子名称: | Envelope glycoprotein gp41 | | 著者 | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | 登録日 | 2016-04-14 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2NCU
 
 | |
2NCV
 
 | |
2NCW
 
 | |
2NCX
 
 | |
2NCY
 
 | |
2NCZ
 
 | |
2ND0
 
 | |
2ND1
 
 | |
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | 分子名称: | De novo mini protein HHH_06 | | 著者 | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | 登録日 | 2016-04-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | 分子名称: | De novo mini protein EEH_04 | | 著者 | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | 登録日 | 2016-04-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND4
 
 | |
2ND5
 
 | | Lysine dimethylated FKBP12 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | 著者 | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | 登録日 | 2016-05-05 | | 公開日 | 2017-05-17 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
2ND6
 
 | | Structure of DK17 in GM1 LUVS | | 分子名称: | Cell penetrating peptide | | 著者 | Bera, S, Bhunia, A. | | 登録日 | 2016-05-11 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND7
 
 | | Structure of DK17 in POPC:POPG:Cholesterol:GM1 LUVS | | 分子名称: | Cell penetrating peptide | | 著者 | Bera, S, Bhunia, A. | | 登録日 | 2016-05-11 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND8
 
 | | Structures of DK17 in TBLE LUVS | | 分子名称: | Cell penetrating peptide | | 著者 | Bera, S, Bhunia, A. | | 登録日 | 2016-05-11 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND9
 
 | | Solution structure of MapZ extracellular domain first subdomain | | 分子名称: | Mid-cell-anchored protein Z | | 著者 | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | 登録日 | 2016-05-11 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2NDA
 
 | | Solution structure of MapZ extracellular domain second subdomain | | 分子名称: | Mid-cell-anchored protein Z | | 著者 | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | 登録日 | 2016-05-11 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2NDB
 
 | |
