5U9A
 
 | |
7W7C
 
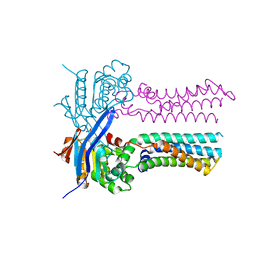 | | Heme exporter in the unliganded form | | 分子名称: | Putative ABC transport system integral membrane protein, Putative ABC transport system, ATP-binding protein, ... | | 著者 | Rahman, M.M, Hisano, T, Nakamura, H, Tosha, T, Shirouzu, M, Shiro, Y. | | 登録日 | 2021-12-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7KIP
 
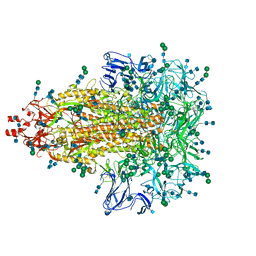 | | A 3.4 Angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Chmielewski, D, Schmid, M, Simmons, G, Jin, J, Chiu, W. | | 登録日 | 2020-10-24 | | 公開日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | A 3.4- angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles.
Biorxiv, 2020
|
|
5U5L
 
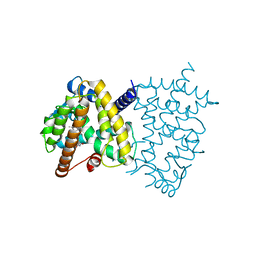 | | X-ray Crystal Structure of the PPARgamma Ligand Binding Domain in Complex with Rivoglitazone | | 分子名称: | (5S)-5-({4-[(6-methoxy-1-methyl-1H-benzimidazol-2-yl)methoxy]phenyl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | 著者 | Bruning, J.B, Rajapaksha, H, Wegener, K, Bhatia, H. | | 登録日 | 2016-12-06 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | X-ray crystal structure of rivoglitazone bound to PPAR gamma and PPAR subtype selectivity of TZDs.
Biochim. Biophys. Acta, 1861, 2017
|
|
5TOI
 
 | |
7W6Z
 
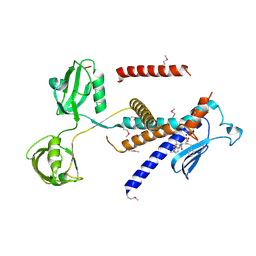 | |
7KQO
 
 | |
7W70
 
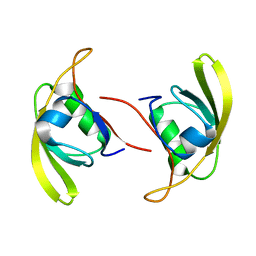 | |
7W6X
 
 | |
7W6Y
 
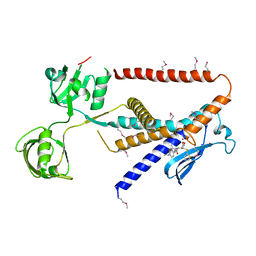 | | Crystal structure of Kangiella koreensis RseP orthologue in complex with batimastat in space group P1 | | 分子名称: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, Anti sigma-E protein, RseA, ... | | 著者 | Imaizumi, Y, Takanuki, K, Nogi, T. | | 登録日 | 2021-12-02 | | 公開日 | 2022-09-07 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Mechanistic insights into intramembrane proteolysis by E. coli site-2 protease homolog RseP.
Sci Adv, 8, 2022
|
|
5UAE
 
 | | Crystal structure of the coiled-coil domain from Listeria Innocua Phage Integrase (Trigonal Form) | | 分子名称: | CITRATE ANION, Putative integrase | | 著者 | Gupta, K, Sharp, R, Yuan, J.B, Van Duyne, G.D. | | 登録日 | 2016-12-19 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Coiled-coil interactions mediate serine integrase directionality.
Nucleic Acids Res., 45, 2017
|
|
5UDO
 
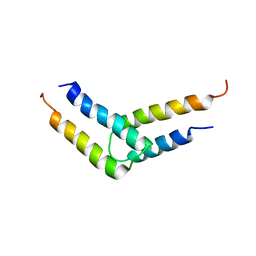 | |
7W6M
 
 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2021-12-02 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
5U9I
 
 | | Crystal structure of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | 分子名称: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | 著者 | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | 登録日 | 2016-12-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7W73
 
 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2021-12-03 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | 分子名称: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-11-28 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
7VZS
 
 | |
5U6R
 
 | |
5U9L
 
 | |
7VZP
 
 | |
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | 分子名称: | CALCIUM ION, Perforin-1 | | 著者 | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | 登録日 | 2017-01-07 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
7W1Q
 
 | |
7L04
 
 | |
5TR5
 
 | |
7W9N
 
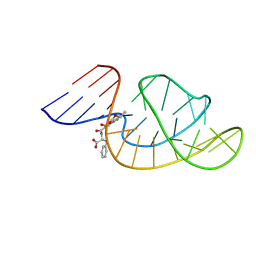 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | 著者 | Xu, G.H, Li, C.G. | | 登録日 | 2021-12-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
