4QRV
 
 | |
8OY0
 
 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Acinetobacter baumanii | | 分子名称: | ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, GLYCEROL, ... | | 著者 | Alphey, M.S, Read, B, da Silva, R.G. | | 登録日 | 2023-05-03 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.
Biochemistry, 63, 2024
|
|
6IX5
 
 | | The structure of LepI complex with SAM and its substrate analogue | | 分子名称: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | 著者 | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | 登録日 | 2018-12-09 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
8ORK
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus | | 分子名称: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | 著者 | De Rose, S.A, Isupov, M. | | 登録日 | 2023-04-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
4EIF
 
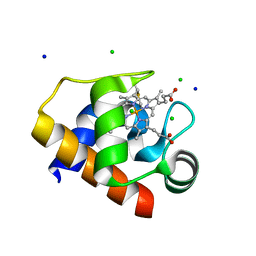 | | Crystal structure of cytochrome c6C L50Q mutant from Synechococcus sp. PCC 7002 | | 分子名称: | CHLORIDE ION, Cytochrome c6, HEME C, ... | | 著者 | Krzywda, S, Bialek, W, Zatwarnicki, P, Jaskolski, M, Szczepaniak, A. | | 登録日 | 2012-04-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Cytochrome c6 and c6C from Synechococcus sp. PCC 7002 - structure and function.
To be Published
|
|
2HEE
 
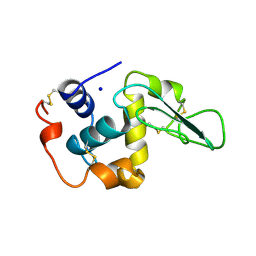 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | 登録日 | 1997-09-16 | | 公開日 | 1998-01-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
4ET8
 
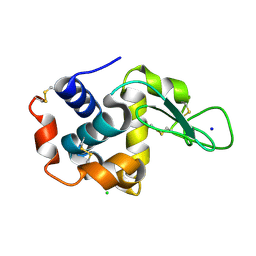 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
8P88
 
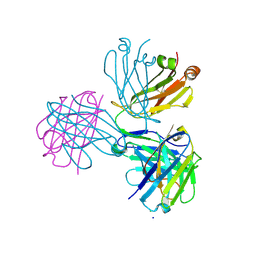 | |
1IVG
 
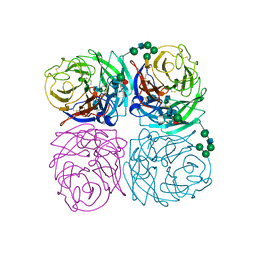 | |
8PH4
 
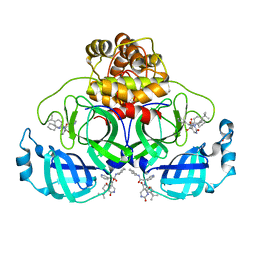 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | 著者 | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | 登録日 | 2023-06-18 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
6J3B
 
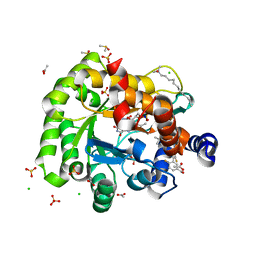 | | Crystal structure of human DHODH in complex with inhibitor 1289 | | 分子名称: | (6R)-1-[3,5-bis(fluoranyl)-4-[2-fluoranyl-5-(hydroxymethyl)phenyl]phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, ACETATE ION, CHLORIDE ION, ... | | 著者 | Yu, Y, Chen, Q. | | 登録日 | 2019-01-04 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
4ADB
 
 | |
8PHA
 
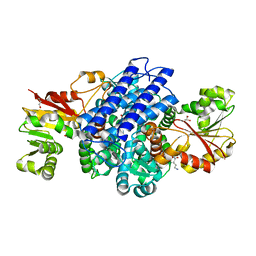 | | O(S)-methyltransferase from Pleurotus sapidus | | 分子名称: | 2-HYDROXY BUTANE-1,4-DIOL, GLYCEROL, L-ornithine, ... | | 著者 | Korf, L, Essen, L.-O. | | 登録日 | 2023-06-19 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | A Novel O - and S -Methyltransferase from Pleurotus sapidus Is Involved in Flavor Formation.
J.Agric.Food Chem., 72, 2024
|
|
8OYH
 
 | | X-ray structure of furin (PCSK3) in complex with Guanidinomethyl-Phac-Can-Tle-Can-6-(aminomethyl)-3-amino-isoindol | | 分子名称: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Dahms, S.O, Brandstetter, H. | | 登録日 | 2023-05-04 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
6JI2
 
 | | Crystal structure of archaeal ribosomal protein aP1, aPelota, and GTP-bound aEF1A complex | | 分子名称: | Archaeal ribosomal stalk protein aP1, Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Maruyama, K, Imai, H, Kawamura, M, Ishino, S, Ishino, Y, Ito, K, Uchiumi, T. | | 登録日 | 2019-02-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Sci Rep, 9, 2019
|
|
4R7C
 
 | | Crystal Structure of CNG mimicking NaK-ETPP mutant cocrystallized with DiMethylammonium | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYLAMINE, GLYCINE, ... | | 著者 | De March, M, Napolitano, L.M.R, Onesti, S. | | 登録日 | 2014-08-27 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2HOH
 
 | | RIBONUCLEASE T1 (N9A MUTANT) COMPLEXED WITH 2'GMP | | 分子名称: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PHOSPHATE ION, ... | | 著者 | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | 登録日 | 1998-09-14 | | 公開日 | 1998-09-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
1IVD
 
 | |
8P58
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer R. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P86
 
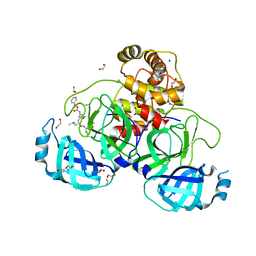 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM MG-132, from an "old" crystal. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-31 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P57
 
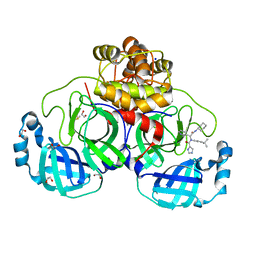 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar X77. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P55
 
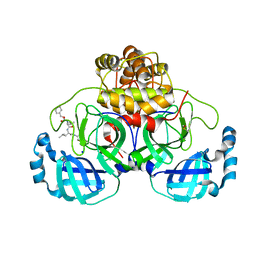 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar MG-132. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5A
 
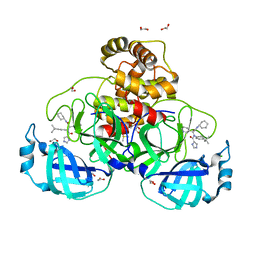 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer R. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P87
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM X77, from an "old" crystal. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-31 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P56
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar X77. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
