4KXP
 
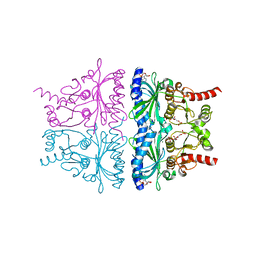 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant I10D in T-state | | 分子名称: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | 著者 | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | 登録日 | 2013-05-27 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mechanism of Displacement of a Catalytically Essential Loop from the Active Site of Mammalian Fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
6BRA
 
 | |
1KRM
 
 | |
3RUB
 
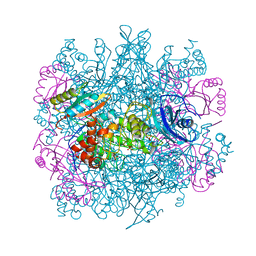 | | CRYSTAL STRUCTURE OF THE UNACTIVATED FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM TOBACCO REFINED AT 2.0-ANGSTROMS RESOLUTION | | 分子名称: | ASPARAGINE, RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, FORM III, ... | | 著者 | Schreuder, H, Cascio, D, Curmi, P.M.G, Chapman, M.S, Suh, S.W, Eisenberg, D.S. | | 登録日 | 1990-05-25 | | 公開日 | 1992-10-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
J.Biol.Chem., 267, 1992
|
|
6C7F
 
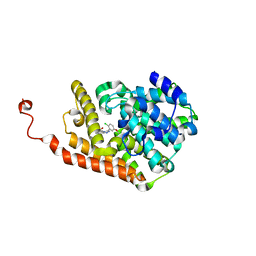 | | Crystal structure of human phosphodiesterase 2A with 1-(2-chloro-5-isobutoxy-phenyl)-N,4-dimethyl-[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide | | 分子名称: | 1-[2-chloro-5-(2-methylpropoxy)phenyl]-N,4-dimethyl[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | 著者 | Xu, R, Aertgeerts, K. | | 登録日 | 2018-01-22 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J. Med. Chem., 61, 2018
|
|
6CK6
 
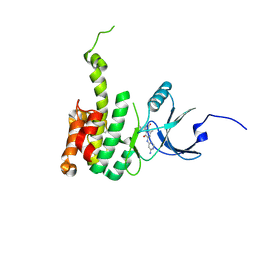 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | 分子名称: | 6'-[(6-aminopyrimidin-4-yl)amino]-8'-methyl-2'H-spiro[cyclohexane-1,3'-imidazo[1,5-a]pyridine]-1',5'-dione, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | 著者 | Han, Q. | | 登録日 | 2018-02-27 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CDR
 
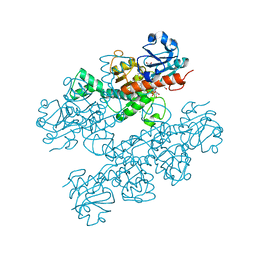 | | Human CtBP1 (28-378) | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | 著者 | Royer, W.E, Bellesis, A.G. | | 登録日 | 2018-02-09 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
8EZO
 
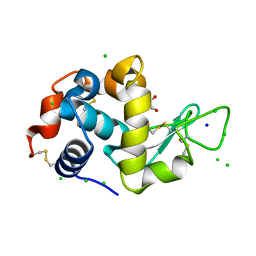 | | Lysozyme Anomalous Dataset at 220 K and 7.1 keV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Doukov, T, Yabukarski, F. | | 登録日 | 2022-11-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6CGR
 
 | |
4L6S
 
 | | PARP complexed with benzo[1,4]oxazin-3-one inhibitor | | 分子名称: | (2S)-6-{[4-(4-chlorophenyl)-3,6-dihydropyridin-1(2H)-yl]methyl}-2-methyl-2H-1,4-benzoxazin-3(4H)-one, Poly [ADP-ribose] polymerase 1 | | 著者 | Dougan, D.R, Mol, C.D, Lawson, J.D. | | 登録日 | 2013-06-12 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of novel benzo[b][1,4]oxazin-3(4H)-ones as poly(ADP-ribose)polymerase inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5ILX
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | 著者 | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2016-03-05 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | 分子名称: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | 著者 | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | 登録日 | 2022-10-24 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
5HSF
 
 | | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1. | | 分子名称: | Ig gamma-1 chain C region, TRIETHYLENE GLYCOL | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Flores, K, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-01-25 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1.
To Be Published
|
|
1Y0M
 
 | |
2RUS
 
 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, MG(II), AND ACTIVATOR CO2 AT 2.3-ANGSTROMS RESOLUTION | | 分子名称: | FORMYL GROUP, MAGNESIUM ION, RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | 著者 | Lundqvist, T, Schneider, G. | | 登録日 | 1991-10-11 | | 公開日 | 1991-10-15 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the ternary complex of ribulose-1,5-bisphosphate carboxylase, Mg(II), and activator CO2 at 2.3-A resolution.
Biochemistry, 30, 1991
|
|
6C7G
 
 | | Crystal structure of human phosphodiesterase 2A with N-(1-adamantyl)-1-(2-chloro-5-isobutoxy-phenyl)-4-methyl-[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide | | 分子名称: | 1-[2-chloro-5-(2-methylpropoxy)phenyl]-4-methyl-N-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl][1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | 著者 | Xu, R, Aertgeerts, K. | | 登録日 | 2018-01-22 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J. Med. Chem., 61, 2018
|
|
8ESY
 
 | | D30N mutant HIV protease in complex with benzoxaborolone analog of darunavir | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Windsor, I.W, Graham, B.J, Raines, R.T. | | 登録日 | 2022-10-15 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
8EJL
 
 | | Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide | | 分子名称: | Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein | | 著者 | Pornillos, O, Ganser-Pornillos, B.K, Schirra, R.T, dos Santos, N.F.B. | | 登録日 | 2022-09-17 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | A molecular switch modulates assembly and host factor binding of the HIV-1 capsid.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EZX
 
 | | Lysozyme Anomalous Dataset at 293 K and 7.1 keV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Doukov, T, Yabukarski, F, Herschlag, D. | | 登録日 | 2022-11-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8EZU
 
 | | Lysozyme Anomalous Dataset at 273 K and 7.1 keV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Doukov, T, Yabukarski, F, Herschlag, D. | | 登録日 | 2022-11-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8EZP
 
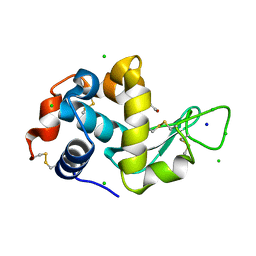 | | Lysozyme Anomalous Dataset at 260 K and 7.1 keV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Doukov, T, Yabukarski, F, Herschlag, D. | | 登録日 | 2022-11-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8F0B
 
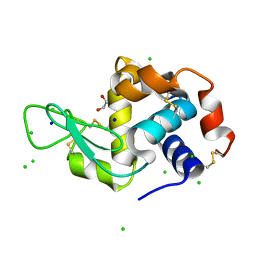 | | Lysozyme Anomalous Dataset at 240 K and 7.1 keV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Doukov, T, Yabukarski, F, Herschlag, D. | | 登録日 | 2022-11-02 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5I8C
 
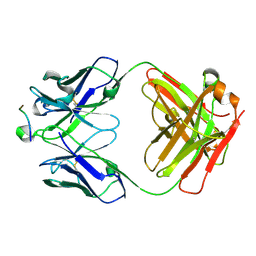 | | Crystal Structure of HIV-1 Clade A BG505 Fusion Peptide (residue 512-520) in Complex with Broadly Neutralizing Antibody VRC34.01 Fab | | 分子名称: | HIV-1 Clade A BG505 Fusion Peptide (residue 512-520), VRC34.01 Fab heavy chain, VRC34.01 Fab light chain | | 著者 | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | 登録日 | 2016-02-18 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
1YI4
 
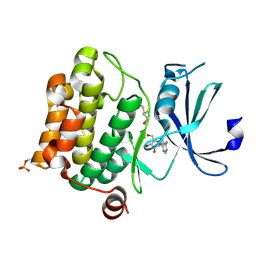 | | Structure of Pim-1 bound to adenosine | | 分子名称: | ADENOSINE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | 著者 | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | 登録日 | 2005-01-11 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
4BZT
 
 | | The Solution Structure of the MLN 944-d(ATGCAT)2 Complex | | 分子名称: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | 著者 | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | 登録日 | 2013-07-30 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
