8ETB
 
 | | the crystal structure of a rationally designed zinc sensor based on maltose binding protein - Zn binding conformation | | 分子名称: | ACETATE ION, ZINC ION, Zinc Sensor protein | | 著者 | Zhao, Z, Zhou, M, Zemerov, S.D, Marmorstein, R, Dmochowski, I.J. | | 登録日 | 2022-10-16 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
5W3Q
 
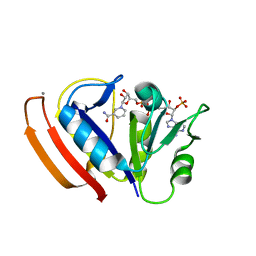 | | L28F E.coli DHFR in complex with NADPH | | 分子名称: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Oyen, D, Wright, P.E, Wilson, I.A. | | 登録日 | 2017-06-08 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
5K6P
 
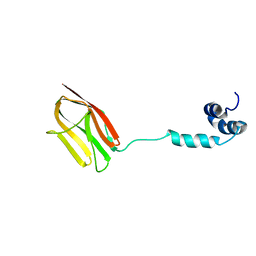 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | 分子名称: | Myosin-binding protein C, cardiac-type | | 著者 | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | 登録日 | 2016-05-25 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | 分子名称: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | 著者 | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | 登録日 | 2019-08-21 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7QGV
 
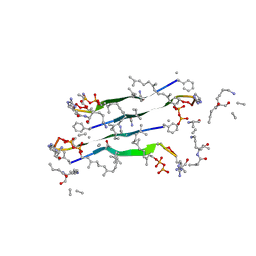 | |
6QFP
 
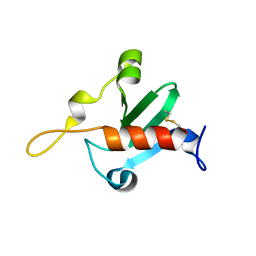 | |
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | 分子名称: | Uncharacterized protein | | 著者 | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
7LQT
 
 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | 分子名称: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | 著者 | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
7MMY
 
 | |
8SD7
 
 | |
8SD1
 
 | |
8SD6
 
 | |
8SD9
 
 | |
8SF1
 
 | |
8SD8
 
 | |
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | 分子名称: | Alpha-crystallin B chain | | 著者 | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | 登録日 | 2011-04-27 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | 主引用文献 | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3KMY
 
 | | Structure of BACE bound to SCH12472 | | 分子名称: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | 分子名称: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KMX
 
 | | Structure of BACE bound to SCH346572 | | 分子名称: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
8GS7
 
 | | SOLUTION NMR STRUCTURE OF N-TERMINAL DOMAIN OF TRICONEPHILA CLAVIPES MAJOR AMPULLATE SPIDROIN 2 | | 分子名称: | Major ampullate spidroin 2 variant 3 | | 著者 | Oktaviani, N.A, Malay, A.D, Matsugami, A, Hayashi, F, Numata, K. | | 登録日 | 2022-09-05 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unusual p K a Values Mediate the Self-Assembly of Spider Dragline Silk Proteins.
Biomacromolecules, 24, 2023
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | 分子名称: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | 著者 | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | 登録日 | 1993-09-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | 分子名称: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | 著者 | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | 登録日 | 1993-09-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
3S9K
 
 | | Crystal structure of the Itk SH2 domain. | | 分子名称: | CITRIC ACID, Tyrosine-protein kinase ITK/TSK | | 著者 | Joseph, R.E, Ginder, N.D, Hoy, J.A, Nix, J.C, Fulton, B.D, Honzatko, R.B, Andreotti, A.H. | | 登録日 | 2011-06-01 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | Structure of the interleukin-2 tyrosine kinase Src homology 2 domain; comparison between X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4YNR
 
 | | DosS GAFA Domain Reduced CO Bound Crystal Structure | | 分子名称: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, Redox sensor histidine kinase response regulator DevS | | 著者 | Madrona, Y. | | 登録日 | 2015-03-10 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Analysis of cytochrome P450 CYP119 ligand-dependent conformational dynamics by two-dimensional NMR and X-ray crystallography.
J.Biol.Chem., 290, 2015
|
|
4YOF
 
 | | DosS GAFA Domain Reduced Nitric Oxide Bound Crystal Structure | | 分子名称: | NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, Redox sensor histidine kinase response regulator DevS | | 著者 | Madrona, Y. | | 登録日 | 2015-03-11 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Analysis of cytochrome P450 CYP119 ligand-dependent conformational dynamics by two-dimensional NMR and X-ray crystallography.
J.Biol.Chem., 290, 2015
|
|
