8CHI
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | 分子名称: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP1A | | 著者 | Meyners, C, Purder, P.L, Hausch, F. | | 登録日 | 2023-02-08 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHK
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | 分子名称: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1A | | 著者 | Meyners, C, Purder, P.L, Hausch, F. | | 登録日 | 2023-02-08 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CQ1
 
 | |
8CPB
 
 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | 分子名称: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | 著者 | Jimenez-Faraco, E, Hermoso, J.A. | | 登録日 | 2023-03-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
8CHJ
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((R)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | 分子名称: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Meyners, C, Purder, P.L, Hausch, F. | | 登録日 | 2023-02-08 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CP9
 
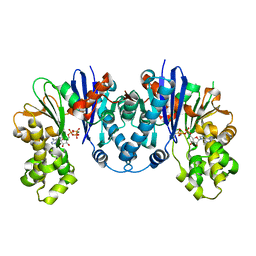 | |
8CPC
 
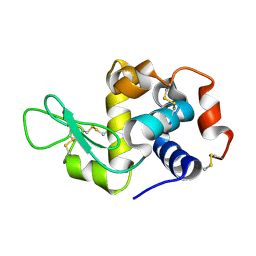 | |
7SHG
 
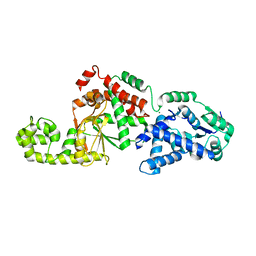 | |
7SKA
 
 | | Sub-tomogram averaged structure of HIV-1 Envelope protein in native membrane | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mangala Prasad, V, Lee, K.K. | | 登録日 | 2021-10-20 | | 公開日 | 2022-03-09 | | 実験手法 | ELECTRON MICROSCOPY (9.1 Å) | | 主引用文献 | Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice.
Cell, 185, 2022
|
|
8CNN
 
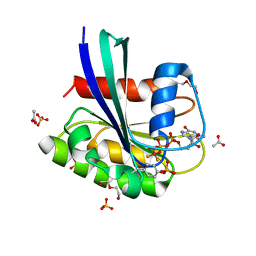 | | BeF3 Phospho-HRas GSA complex | | 分子名称: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Baumann, P, Jin, Y. | | 登録日 | 2023-02-23 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
8CNJ
 
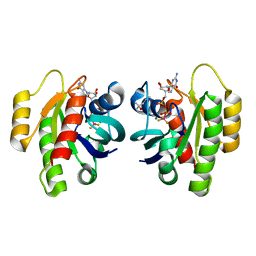 | | HRas(1-166) in complex with GDP and BeF3- | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, BERYLLIUM TRIFLUORIDE ION, GTPase HRas, ... | | 著者 | Baumann, P, Jin, Y. | | 登録日 | 2023-02-23 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
8CQM
 
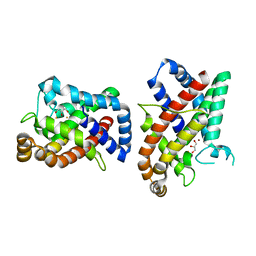 | |
7SCP
 
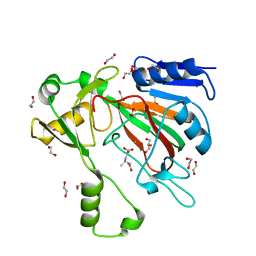 | | The crystal structure of ScoE in complex with intermediate | | 分子名称: | (3R)-3-(oxaloamino)butanoic acid, 1,2-ETHANEDIOL, FE (II) ION, ... | | 著者 | Cha, L, Chen, J, Zhou, J, Chang, W. | | 登録日 | 2021-09-28 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Deciphering the Reaction Pathway of Mononuclear Iron Enzyme-Catalyzed N-C Triple Bond Formation in Isocyanide Lipopeptide and Polyketide Biosynthesis
Acs Catalysis, 12, 2022
|
|
1PZR
 
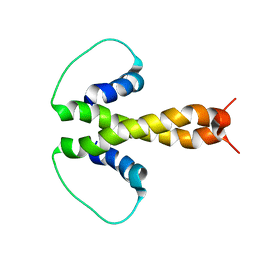 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | 分子名称: | Erythronolide synthase | | 著者 | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | 登録日 | 2003-07-14 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1PZQ
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS 2 and DEBS 3: The A domain | | 分子名称: | Erythronolide synthase | | 著者 | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | 登録日 | 2003-07-14 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1SO9
 
 | | Solution Structure of apoCox11, 30 structures | | 分子名称: | Cytochrome C oxidase assembly protein ctaG | | 著者 | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-03-13 | | 公開日 | 2004-08-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1PQT
 
 | |
1SZO
 
 | | Crystal Structure Analysis of the 6-Oxo Camphor Hydrolase His122Ala Mutant Bound to Its Natural Product (2S,4S)-alpha-Campholinic Acid | | 分子名称: | (2S,4S)-4-(2,2-DIHYDROXYETHYL)-2,3,3-TRIMETHYLCYCLOPENTANONE, 6-oxocamphor hydrolase, CALCIUM ION | | 著者 | Leonard, P.M, Grogan, G. | | 登録日 | 2004-04-06 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of 6-oxo camphor hydrolase H122A mutant bound to its natural product, (2S,4S)-alpha-campholinic acid: mutant structure suggests an atypical mode of transition state binding for a crotonase homolog.
J.Biol.Chem., 279, 2004
|
|
1RWD
 
 | |
1P3H
 
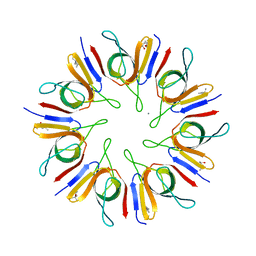 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | 著者 | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2003-04-17 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
1T5A
 
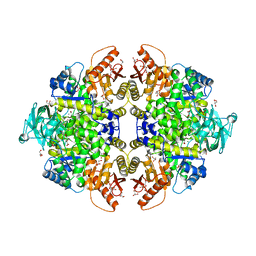 | | Human Pyruvate Kinase M2 | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Dombrauckas, J.D, Santarsiero, B.D, Mesecar, A.D. | | 登録日 | 2004-05-03 | | 公開日 | 2005-07-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis.
Biochemistry, 44, 2005
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | 分子名称: | Cytochrome C oxidase assembly protein ctaG | | 著者 | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | 登録日 | 2004-03-16 | | 公開日 | 2004-08-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
5AZ2
 
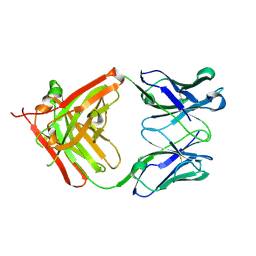 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | 分子名称: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | 著者 | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | 登録日 | 2015-09-16 | | 公開日 | 2015-12-09 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.603 Å) | | 主引用文献 | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5B4N
 
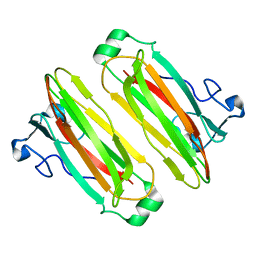 | | Structure analysis of function associated loop mutant of substrate recognition domain of Fbs1 ubiquitin ligase | | 分子名称: | F-box only protein 2 | | 著者 | Nishio, K, Yoshida, Y, Tanaka, K, Mizushima, T. | | 登録日 | 2016-04-06 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural analysis of a function-associated loop mutant of the substrate-recognition domain of Fbs1 ubiquitin ligase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B62
 
 | | Crystal structure of N-terminal amidase with Asn-Glu-Ala peptide | | 分子名称: | ASN-GLU-ALA, Nta1p | | 著者 | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | 登録日 | 2016-05-24 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.042 Å) | | 主引用文献 | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
