7TJJ
 
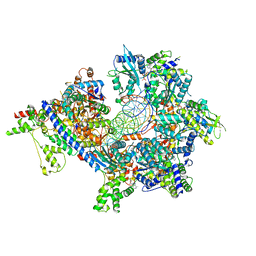 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
6W0M
 
 | |
6W0R
 
 | |
5MHJ
 
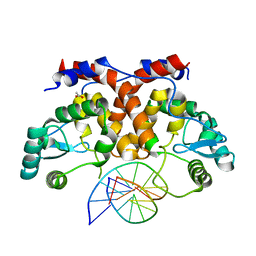 | | ICP4 DNA-binding domain, lacking intrinsically disordered region, in complex with 12mer DNA duplex from its own promoter | | 分子名称: | ACETIC ACID, CHLORIDE ION, DNA (5'-D(P*CP*GP*AP*TP*CP*GP*TP*CP*C)-3'), ... | | 著者 | Tunnicliffe, R.B, Lockhart-Cairns, M.P, Levy, C, Mould, P, Jowitt, T.A, Sito, H, Baldock, C, Sandri-Goldin, R.M, Golovanov, A.P. | | 登録日 | 2016-11-24 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.117 Å) | | 主引用文献 | The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res., 45, 2017
|
|
7TJI
 
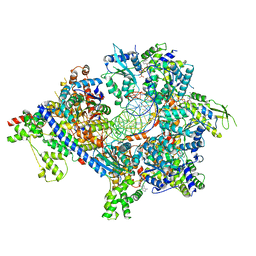 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with flexible Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJH
 
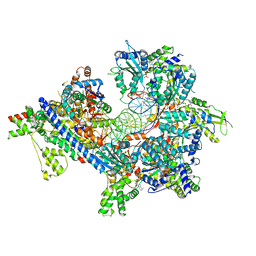 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with flexible Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
2YG9
 
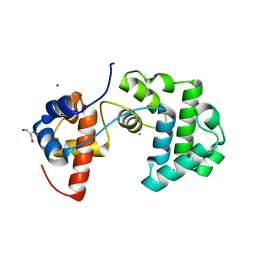 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | 分子名称: | CHLORIDE ION, DNA-3-methyladenine glycosidase II, putative, ... | | 著者 | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | 登録日 | 2011-04-11 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
1U8R
 
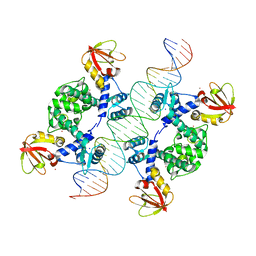 | | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions | | 分子名称: | COBALT (II) ION, Iron-dependent repressor ideR, SODIUM ION, ... | | 著者 | Wisedchaisri, G, Holmes, R.K, Hol, W.G.J. | | 登録日 | 2004-08-06 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions.
J.Mol.Biol., 342, 2004
|
|
7B24
 
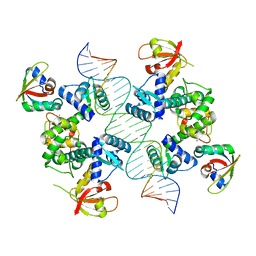 | |
7B1Y
 
 | |
7B23
 
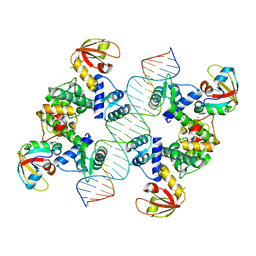 | |
7B20
 
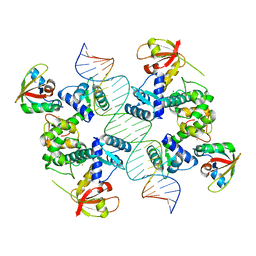 | |
7B25
 
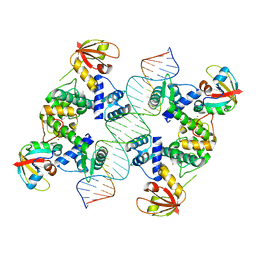 | |
4GFB
 
 | | Rap1/DNA complex | | 分子名称: | CALCIUM ION, DNA-binding protein RAP1, telomeric DNA | | 著者 | Le Bihan, Y.-V, Matot, B, Le Du, M.-H. | | 登録日 | 2012-08-03 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Effect of Rap1 binding on DNA distortion and potassium permanganate hypersensitivity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1MSE
 
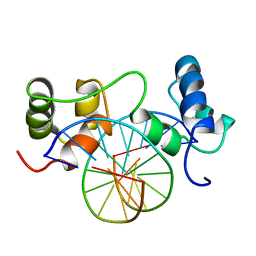 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | 分子名称: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-01-24 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
1MSF
 
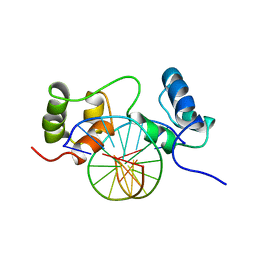 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | 分子名称: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-01-24 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
2WWY
 
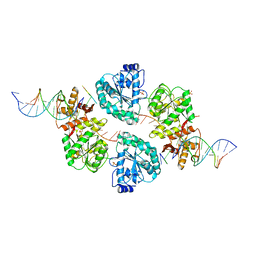 | | Structure of human RECQ-like helicase in complex with a DNA substrate | | 分子名称: | 1,2-ETHANEDIOL, 5'-D(*DA DG DC DG DT DC DG DA DG DA DT DC DCP)-3', ATP-DEPENDENT DNA HELICASE Q1, ... | | 著者 | Pike, A.C.W, Zhang, Y, Schnecke, C, Chaikuad, A, Krojer, T, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2009-10-30 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Recq1 Helicase-Driven DNA Unwinding, Annealing, and Branch Migration: Insights from DNA Complex Structures
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5K5Q
 
 | |
4HQB
 
 | |
4IUF
 
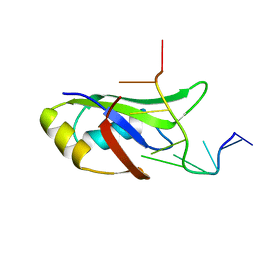 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with a Single-stranded DNA | | 分子名称: | 5'-D(*GP*TP*TP*GP*(XUA)P*GP*CP*GP*T)-3', TAR DNA-binding protein 43 | | 著者 | Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yang, W.Z, Yuan, H.S. | | 登録日 | 2013-01-21 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.752 Å) | | 主引用文献 | The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids.
Nucleic Acids Res., 42, 2014
|
|
4C8M
 
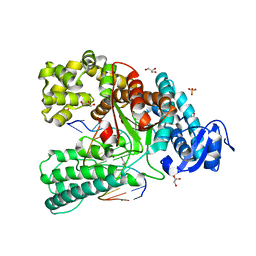 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair d5SICS-dNaM at the postinsertion site (sequence context 2) | | 分子名称: | GLYCEROL, LARGE FRAGMENT OF TAQ DNA POLYMERASE I, MAGNESIUM ION, ... | | 著者 | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | 登録日 | 2013-10-01 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.568 Å) | | 主引用文献 | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8N
 
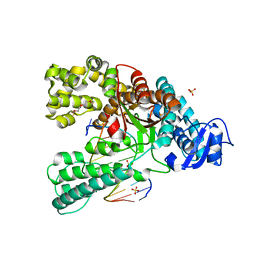 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 3) | | 分子名称: | LARGE FRAGMENT OF TAQ DNA POLYMERASE I, PRIMER, 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', ... | | 著者 | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | 登録日 | 2013-10-01 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8L
 
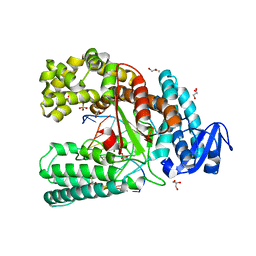 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the artificial base pair dNaM-d5SICS at the postinsertion site (sequence context 1) | | 分子名称: | 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*AP*GP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP)-3', DNA POLYMERASE I, ... | | 著者 | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | 登録日 | 2013-10-01 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | 著者 | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | 登録日 | 2022-01-13 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
4C8O
 
 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 2) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*TP*TP*CP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*CP)-3', ... | | 著者 | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | 登録日 | 2013-10-01 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
