3B36
 
 | | Structure of M26L DJ-1 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein DJ-1 | | 著者 | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | 登録日 | 2007-10-19 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
3B3A
 
 | | Structure of E163K/R145E DJ-1 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein DJ-1 | | 著者 | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | 登録日 | 2007-10-19 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | 分子名称: | CRISPR-associated protein Cas1 | | 著者 | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | 登録日 | 2010-06-18 | | 公開日 | 2010-08-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
3LYT
 
 | |
3B58
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38G Mutation and a 2',5'-Phosphodiester Linkage at the Active Site | | 分子名称: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2007-10-25 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | 分子名称: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | 著者 | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | 登録日 | 2010-03-10 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3PCD
 
 | | PROTOCATECHUATE 3,4-DIOXYGENASE Y447H MUTANT | | 分子名称: | BETA-MERCAPTOETHANOL, CARBONATE ION, FE (III) ION, ... | | 著者 | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | 登録日 | 1997-11-24 | | 公開日 | 1998-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
Biochemistry, 37, 1998
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | 分子名称: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | 著者 | Sacco, M, Wang, J, Chen, Y. | | 登録日 | 2020-12-03 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
5OGL
 
 | | Structure of bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog | | 分子名称: | MANGANESE (II) ION, SODIUM ION, Substrate mimicking peptide, ... | | 著者 | Napiorkowska, M, Boilevin, J, Sovdat, T, Darbre, T, Reymond, J.-L, Aebi, M, Locher, K.P. | | 登録日 | 2017-07-13 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OD5
 
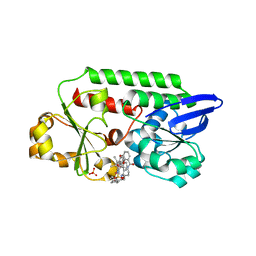 | | Periplasmic binding protein CeuE complexed with a synthetic catalyst | | 分子名称: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 4-(aminomethyl)-~{N}-(pyridin-2-ylmethyl)benzenesulfonamide, Azotochelin, ... | | 著者 | Duhme-Klair, A.K, Raines, D.J, Clarke, J.E, Blagova, E.V, Dodson, E.J, Wilson, K.S. | | 登録日 | 2017-07-04 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
5NBD
 
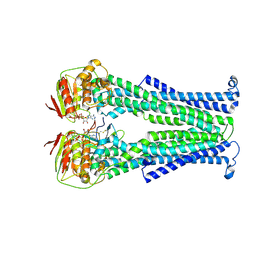 | | PglK flippase in complex with inhibitory nanobody | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | 著者 | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | 登録日 | 2017-03-01 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
4GPD
 
 | |
4FX2
 
 | |
3AT1
 
 | |
5MDH
 
 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF PORCINE CYTOPLASMIC MALATE DEHYDROGENASE ALPHA-KETOMALONATE AND TNAD AT 2.4 ANGSTROMS RESOLUTION | | 分子名称: | ALPHA-KETOMALONIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Chapman, A.D.M, Cortes, A, Dafforn, T.R, Clarke, A.R, Brady, R.L. | | 登録日 | 1998-10-08 | | 公開日 | 1999-05-18 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD.
J.Mol.Biol., 285, 1999
|
|
3FX2
 
 | |
1KBN
 
 | | Glutathione transferase mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLYCEROL, ... | | 著者 | Rossjohn, J, Parker, M.W. | | 登録日 | 2001-11-06 | | 公開日 | 2003-11-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Glutathione transferase mutant
To be Published
|
|
5K4C
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 2 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit D, GLYCEROL | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4B
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | 分子名称: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4D
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit D | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
4AWI
 
 | | Human Jnk1alpha kinase with 4-phenyl-7-azaindole IKK2 inhibitor. | | 分子名称: | MITOGEN-ACTIVATED PROTEIN KINASE 8, N-(1,1-dioxidotetrahydro-2H-thiopyran-4-yl)-4-[2-(1-methylethyl)-1H-pyrrolo[2,3-b]pyridin-4-yl]benzenesulfonamide, SULFATE ION | | 著者 | Chung, C, Vicentini, G, Liddle, J, Bamborough, P. | | 登録日 | 2012-06-03 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | 4-Phenyl-7-Azaindoles as Potent, Selective and Bioavailable Ikk2 Inhibitors Demonstrating Good in Vivo Efficacy.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
9GSS
 
 | |
1EOG
 
 | | CRYSTAL STRUCTURE OF PI CLASS GLUTATHIONE TRANSFERASE | | 分子名称: | GLUTATHIONE S-TRANSFERASE | | 著者 | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | 登録日 | 2000-03-22 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
1EOH
 
 | | GLUTATHIONE TRANSFERASE P1-1 | | 分子名称: | GLUTATHIONE S-TRANSFERASE | | 著者 | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | 登録日 | 2000-03-22 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
