7M3I
 
 | |
5MOH
 
 | | Crystal structure of CK2alpha with ZT0583 bound. | | 分子名称: | 2-(3-methoxy-4-oxidanyl-phenyl)ethanoic acid, ACETATE ION, Casein kinase II subunit alpha | | 著者 | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | 登録日 | 2016-12-14 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5MWC
 
 | | Crystal structure of the genetically-encoded green calcium indicator NTnC in its calcium bound state | | 分子名称: | CALCIUM ION, genetically-encoded green calcium indicator NTnC | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Rakitina, T.V, Popov, V.O, Subach, O.M, Barykina, N.V, Subach, F.V. | | 登録日 | 2017-01-18 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Enchanced variant of genetically-encoded green calcium indicator NTnC
To Be Published
|
|
3JUE
 
 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | 分子名称: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | 著者 | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | 登録日 | 2009-09-15 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
2YZD
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with 8-azaxanthin (inhibitor) | | 分子名称: | 8-AZAXANTHINE, Uricase | | 著者 | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | 登録日 | 2007-05-05 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
7GAV
 
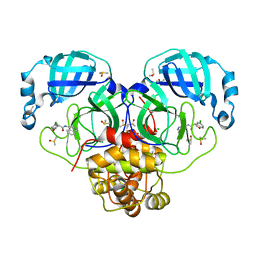 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-f7918075-2 (SARS2_MproA-x0854) | | 分子名称: | (3S)-5-chloro-N-(isoquinolin-4-yl)-2,3-dihydro-1-benzofuran-3-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5MPI
 
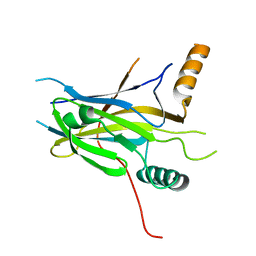 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | 分子名称: | Grainyhead-like protein 1 homolog | | 著者 | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | 登録日 | 2016-12-16 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.345 Å) | | 主引用文献 | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
3JWV
 
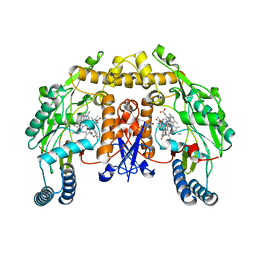 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'S,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JX8
 
 | |
2PLV
 
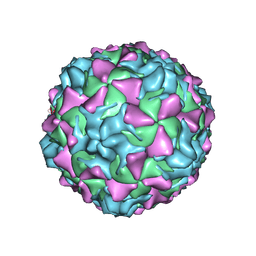 | |
3JYR
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltose-binding periplasmic protein | | 著者 | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | 登録日 | 2009-09-22 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
2G59
 
 | |
5MX8
 
 | |
5MXB
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin | | 分子名称: | Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, SODIUM ION, ... | | 著者 | Sliwiak, J, Sikorski, M, Jaskolski, M. | | 登録日 | 2017-01-22 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
7GAW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e194df51-1 (SARS2_MproA-x0862) | | 分子名称: | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.812 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2G5P
 
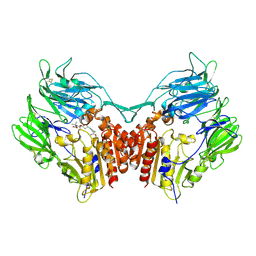 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ac | | 分子名称: | 4-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-3-TERT-BUTYLBENZOIC ACID, Dipeptidyl peptidase 4 | | 著者 | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | 登録日 | 2006-02-23 | | 公開日 | 2006-07-04 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
3JZU
 
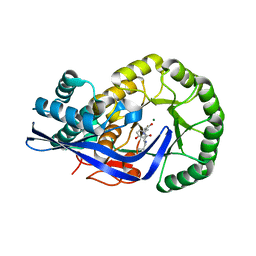 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | 分子名称: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-09-24 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | 分子名称: | IODIDE ION, Protein Ten1 | | 著者 | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | 登録日 | 2009-09-25 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K11
 
 | |
3JDW
 
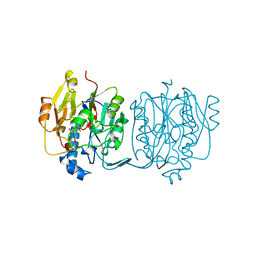 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | 分子名称: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | 著者 | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | 登録日 | 1997-01-24 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
2PSN
 
 | |
3KK6
 
 | | Crystal Structure of Cyclooxygenase-1 in complex with celecoxib | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, CITRATE ANION, ... | | 著者 | Sidhu, R.S. | | 登録日 | 2009-11-04 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5L6D
 
 | |
7M2W
 
 | | Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | 著者 | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | 登録日 | 2021-03-17 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7GBR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-7e92b6ca-4 (Mpro-x10403) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(2-anilinoethyl)-2-oxo-1,2-dihydroquinoline-4-carboxamide | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
