1ZOW
 
 | | Crystal Structure of S. aureus FabH, beta-ketoacyl carrier protein synthase III | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase III | | 著者 | Qiu, X, Choudhry, A.E, Janson, C.A, Grooms, M, Daines, R.A, Lonsdale, J.T, Khandekar, S.S. | | 登録日 | 2005-05-15 | | 公開日 | 2005-08-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus.
Protein Sci., 14, 2005
|
|
4S3C
 
 | | IspG in complex with Epoxide Intermediate | | 分子名称: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | 著者 | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | 登録日 | 2015-01-26 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4P1M
 
 | |
3E5P
 
 | | Crystal structure of alanine racemase from E.faecalis | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine racemase, NONAETHYLENE GLYCOL, ... | | 著者 | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | 登録日 | 2008-08-14 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
2WMP
 
 | | Structure of the E. coli chaperone PapD in complex with the pilin domain of the PapGII adhesin | | 分子名称: | CHAPERONE PROTEIN PAPD, PAPG PROTEIN | | 著者 | Ford, B.A, Verger, D, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | 登録日 | 2009-07-02 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Papd-Papgii Pilin Complex Reveals an Open and Flexible P5 Pocket.
J.Bacteriol., 194, 2012
|
|
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | 分子名称: | GREEN FLUORESCENT PROTEIN | | 著者 | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | 登録日 | 2001-06-07 | | 公開日 | 2003-01-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
5WHS
 
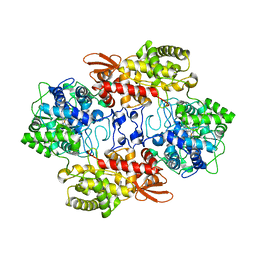 | | Crystal structure of the catalase-peroxidase from Neurospora crassa at 2.6 A | | 分子名称: | Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Diaz-Vilchis, A, Vega-Garcia, V, Rudino-Pinera, E, Hansberg, W. | | 登録日 | 2017-07-18 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure, kinetics, molecular and redox properties of a cytosolic and developmentally regulated fungal catalase-peroxidase.
Arch. Biochem. Biophys., 640, 2018
|
|
1GER
 
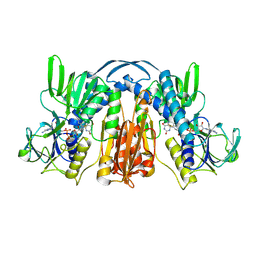 | |
5AGX
 
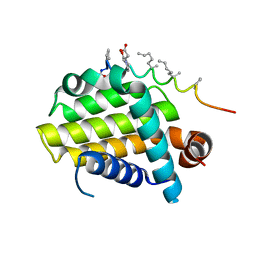 | | Bcl-2 alpha beta-1 LINEAR complex | | 分子名称: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | 著者 | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
1Y53
 
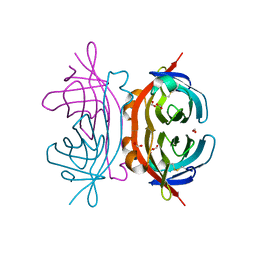 | | Crystal structure of bacterial expressed avidin related protein 4 (AVR4) C122S | | 分子名称: | Avidin-related protein 4/5, FORMIC ACID | | 著者 | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | 登録日 | 2004-12-02 | | 公開日 | 2005-05-24 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2VV5
 
 | |
1LB2
 
 | | Structure of the E. coli alpha C-terminal domain of RNA polymerase in complex with CAP and DNA | | 分子名称: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Benoff, B, Yang, H, Lawson, C.L, Parkinson, G, Liu, J, Blatter, E, Ebright, Y.W, Berman, H.M, Ebright, R.H. | | 登録日 | 2002-04-01 | | 公開日 | 2002-09-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis of transcription activation: the CAP-alpha CTD-DNA complex.
Science, 297, 2002
|
|
1L6S
 
 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4,7-Dioxosebacic Acid | | 分子名称: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | 著者 | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | 登録日 | 2002-03-13 | | 公開日 | 2002-04-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
4S3E
 
 | | IspG in complex with Inhibitor 7 (compound 1061) | | 分子名称: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | 登録日 | 2015-01-26 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
1ZS3
 
 | | The crystal structure of the Lactococcus lactis MG1363 DpsB protein | | 分子名称: | Lactococcus lactis MG1363 DpsA | | 著者 | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | 登録日 | 2005-05-23 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
5YWY
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | 分子名称: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | 著者 | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | 登録日 | 2017-11-30 | | 公開日 | 2018-12-05 | | 最終更新日 | 2018-12-19 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
1KN9
 
 | |
1UMP
 
 | |
5WHQ
 
 | | Crystal structure of the catalase-peroxidase from Neurospora crassa at 2.9 A | | 分子名称: | Catalase-peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Diaz-Vilchis, A, Vega-Garcia, V, Rudino-Pinera, E, Hansberg, W. | | 登録日 | 2017-07-18 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure, kinetics, molecular and redox properties of a cytosolic and developmentally regulated fungal catalase-peroxidase.
Arch. Biochem. Biophys., 640, 2018
|
|
4KMQ
 
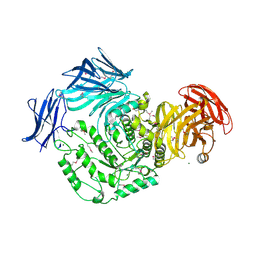 | | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e | | 分子名称: | Lmo2446 protein, MAGNESIUM ION | | 著者 | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-05-08 | | 公開日 | 2013-05-29 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure to function of an alpha-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis.
Nat Microbiol, 2, 2016
|
|
5TKV
 
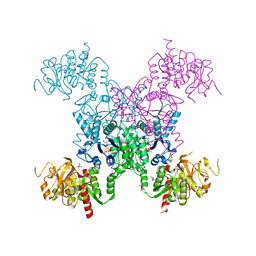 | | X-RAY CRYSTAL STRUCTURE OF THE "CLOSED" CONFORMATION OF CTP-INHIBITED E. COLI CYTIDINE TRIPHOSPHATE (CTP) SYNTHETASE | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, ... | | 著者 | Baldwin, E.P, Endrizzi, J.A. | | 登録日 | 2016-10-07 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4S38
 
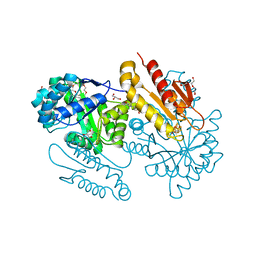 | | IspG in complex MEcPP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | 著者 | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | 登録日 | 2015-01-26 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
1L6Y
 
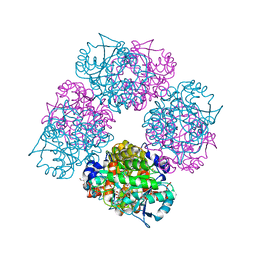 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4-Oxosebacic Acid | | 分子名称: | 4-OXODECANEDIOIC ACID, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | 登録日 | 2002-03-14 | | 公開日 | 2002-04-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
1USL
 
 | | Structure Of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, Complexed With Phosphate. | | 分子名称: | PHOSPHATE ION, RIBOSE 5-PHOSPHATE ISOMERASE B | | 著者 | Roos, A.K, Andersson, C.E, Unge, T, Jones, T.A, Mowbray, S.L. | | 登録日 | 2003-11-25 | | 公開日 | 2004-01-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase Has a Known Fold, But a Novel Active Site
J.Mol.Biol., 335, 2004
|
|
3C8E
 
 | | Crystal Structure Analysis of yghU from E. Coli | | 分子名称: | GLUTATHIONE, yghU, glutathione S-transferase homologue | | 著者 | Harp, J, Ladner, J.E, Schaab, M.R, Stourman, N.V, Armstrong, R.N. | | 登録日 | 2008-02-11 | | 公開日 | 2009-02-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Function of YghU, a Nu-Class Glutathione Transferase Related to YfcG from Escherichia coli.
Biochemistry, 50, 2011
|
|
