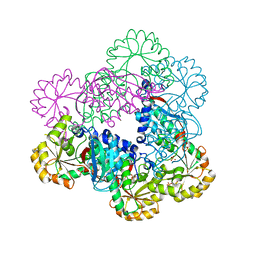
4K4R
 
 | |
5GIH
 
 | |
3KMY
 
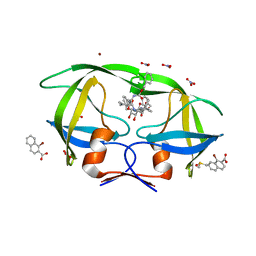
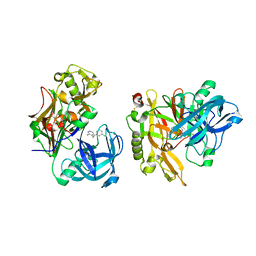 | | Structure of BACE bound to SCH12472 | | 分子名称: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
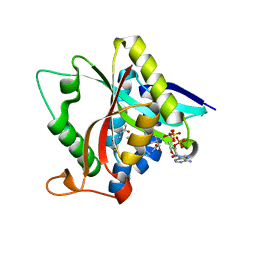
5HUP
 
 | | Crystal Structure of NadC from Streptococcus pyogenes | | 分子名称: | Nicotinate-nucleotide pyrophosphorylase (Carboxylating), SULFATE ION | | 著者 | Booth, W.T, Chruszcz, M. | | 登録日 | 2016-01-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
3KMX
 
 | | Structure of BACE bound to SCH346572 | | 分子名称: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | 分子名称: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
5FHQ
 
 | |
3MNP
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, V708A, E711G) mutations at 1.50A | | 分子名称: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | 著者 | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | 登録日 | 2010-04-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3MNE
 
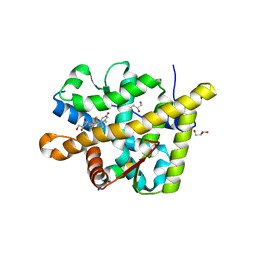 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by F608S mutation at 1.96A | | 分子名称: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | 著者 | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | 登録日 | 2010-04-21 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
1C02
 
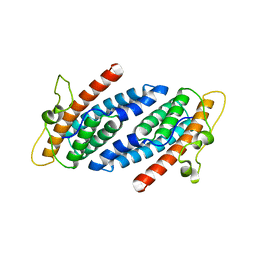 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | 分子名称: | PHOSPHOTRANSFERASE YPD1P | | 著者 | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | 登録日 | 1999-07-14 | | 公開日 | 2000-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
3M8E
 
 | |
1NQA
 
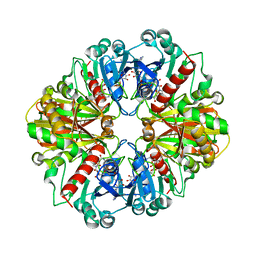 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | 登録日 | 2003-01-21 | | 公開日 | 2003-04-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
5DQ0
 
 | | Structure of human neuropilin-2 b1 domain with novel and unique zinc binding site | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Tsai, Y.I, Rana, R.R, Zachary, I, Djordjevic, S. | | 登録日 | 2015-09-14 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of human neuropilin-2 b1 domain with novel and unique zinc binding site
To Be Published
|
|
3M8K
 
 | |
3M89
 
 | | Structure of TubZ-GTP-g-S | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, FtsZ/tubulin-related protein | | 著者 | Ni, L, Xu, W, Schumacher, M.A. | | 登録日 | 2010-03-17 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M8F
 
 | |
7CVN
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X, Zhou, J, Xu, B. | | 登録日 | 2020-08-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
4K4P
 
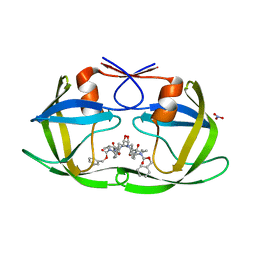 | | TL-3 inhibited Trp6Ala HIV Protease | | 分子名称: | HIV-1 protease, NITRATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | 著者 | Tiefenbrunn, T, Stout, C.D. | | 登録日 | 2013-04-12 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystallographic Fragment-Based Drug Discovery: Use of a Brominated Fragment Library Targeting HIV Protease.
Chem.Biol.Drug Des., 83, 2014
|
|
1OFI
 
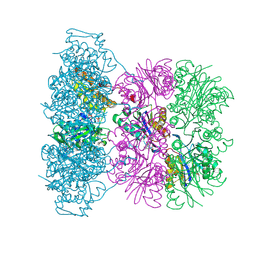 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | 分子名称: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | 著者 | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2003-04-14 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1NQ5
 
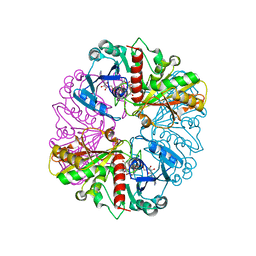 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | 分子名称: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | 登録日 | 2003-01-21 | | 公開日 | 2003-04-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPT
 
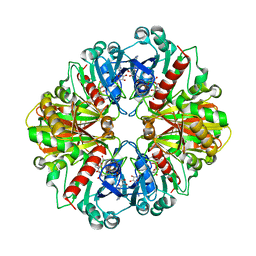 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | 分子名称: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | 登録日 | 2003-01-20 | | 公開日 | 2003-04-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1OFH
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | 著者 | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2003-04-14 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
3TGR
 
 | |
1K58
 
 | | Crystal Structure of Human Angiogenin Variant D116H | | 分子名称: | Angiogenin | | 著者 | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | 登録日 | 2001-10-10 | | 公開日 | 2002-03-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | 著者 | Kwon, Y.D, Kwong, P.D. | | 登録日 | 2011-08-17 | | 公開日 | 2012-04-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
