1HFY
 
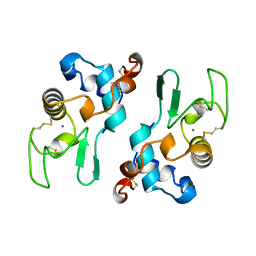 | | ALPHA-LACTALBUMIN | | 分子名称: | ALPHA-LACTALBUMIN, CALCIUM ION | | 著者 | Pike, A.C.W, Brew, K, Acharya, K.R. | | 登録日 | 1996-06-13 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase.
Structure, 4, 1996
|
|
7S4E
 
 | |
1GT0
 
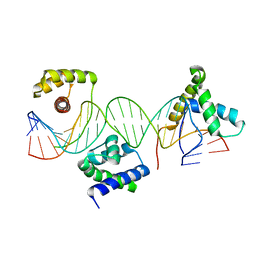 | | Crystal structure of a POU/HMG/DNA ternary complex | | 分子名称: | 5'-D(*AP*TP*CP*CP*CP*AP*TP*TP*AP*GP* CP*AP*TP*CP*CP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', 5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*TP*TP* GP*GP*AP* TP*GP*CP*TP*AP*AP*TP*GP*GP*GP*A)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1, ... | | 著者 | Remenyi, A, Wilmanns, M. | | 登録日 | 2002-01-09 | | 公開日 | 2003-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of a POU/Hmg/DNA Ternary Complex Suggests Differential Assembly of Oct4 and Sox2 on Two Enhancers
Genes Dev., 17, 2003
|
|
7QPU
 
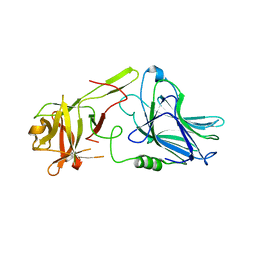 | | Botulinum neurotoxin A5 cell binding domain in complex with GM1b oligosaccharide | | 分子名称: | Botulinum neurotoxin sub-type A5, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | 著者 | Gregory, K.S, Acharya, K.R, Liu, S.M. | | 登録日 | 2022-01-05 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
7QPT
 
 | | Botulinum neurotoxin A4 cell binding domain in complex with GD1a oligosaccharide | | 分子名称: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type A | | 著者 | Gregory, K.S, Acharya, K.R, Liu, S.M, Mojanaga, O.O. | | 登録日 | 2022-01-05 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
7TEI
 
 | |
7TF8
 
 | |
5NDJ
 
 | | Crystal structure of aminoglycoside TC007 in complex with 70S ribosome from Thermus thermophilus, three tRNAs and mRNA (soaking) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | 登録日 | 2017-03-08 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1XZ2
 
 | | wild-type hemoglobin deoxy no-salt | | 分子名称: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kavanaugh, J.S, Rogers, P.H, Arnone, A, Hui, H.L, Wierzba, A, DeYoung, A, Kwiatkowski, L.D, Noble, R.W, Juszczak, L.J, Peterson, E.S, Friedman, J.M. | | 登録日 | 2004-11-11 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Intersubunit interactions associated with tyr42alpha stabilize the quaternary-T tetramer but are not major quaternary constraints in deoxyhemoglobin
Biochemistry, 44, 2005
|
|
8JOK
 
 | |
8JQW
 
 | |
8JNU
 
 | |
3MNO
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, F608S) mutations at 1.55A | | 分子名称: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | 著者 | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | 登録日 | 2010-04-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
5NDK
 
 | | Crystal structure of aminoglycoside TC007 co-crystallized with 70S ribosome from Thermus thermophilus, three tRNAs and mRNA | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | 登録日 | 2017-03-08 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3MNP
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, V708A, E711G) mutations at 1.50A | | 分子名称: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | 著者 | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | 登録日 | 2010-04-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
6ZPU
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain with inserted symmetry molecule C-terminus. | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Cozier, G.E, Acharya, K.R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Angiotensin-converting enzyme open for business: structural insights into the subdomain dynamics.
Febs J., 288, 2021
|
|
5JHY
 
 | |
5JHZ
 
 | |
6ZPT
 
 | |
6ZPQ
 
 | |
4L4R
 
 | | Structural Characterisation of the Apo-form of Human Lactate Dehydrogenase M Isozyme | | 分子名称: | L-lactate dehydrogenase A chain | | 著者 | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | 登録日 | 2013-06-09 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3M8E
 
 | |
3MNE
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by F608S mutation at 1.96A | | 分子名称: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | 著者 | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | 登録日 | 2010-04-21 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | 分子名称: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KMX
 
 | | Structure of BACE bound to SCH346572 | | 分子名称: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
