1VEA
 
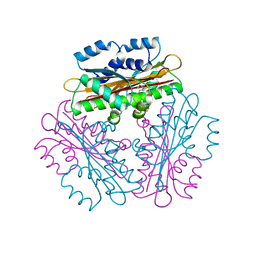 | | Crystal Structure of HutP, an RNA binding antitermination protein | | 分子名称: | Hut operon positive regulatory protein, N-(2-NAPHTHYL)HISTIDINAMIDE | | 著者 | Kumarevel, T.S, Fujimoto, Z, Karthe, P, Oda, M, Mizuno, H, Kumar, P.K.R. | | 登録日 | 2004-03-29 | | 公開日 | 2004-07-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Activated HutP; An RNA Binding Protein that Regulates Transcription of the hut Operon in Bacillus subtilis
Structure, 12, 2004
|
|
1SNY
 
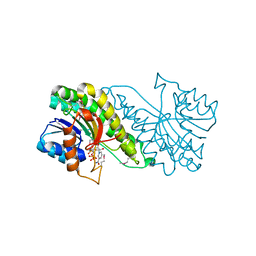 | | Carbonyl reductase Sniffer of D. melanogaster | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sniffer CG10964-PA | | 著者 | Sgraja, T, Ulschmid, J, Becker, K, Schneuwly, S, Klebe, G, Reuter, K, Heine, A. | | 登録日 | 2004-03-12 | | 公開日 | 2004-09-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Insights into the Neuroprotective-acting Carbonyl Reductase Sniffer of Drosophila melanogaster.
J.Mol.Biol., 342, 2004
|
|
3DD4
 
 | |
6QIO
 
 | |
5DZO
 
 | |
7WZO
 
 | |
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2012-05-02 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7X39
 
 | | Structure of CIZ1 bound ERH | | 分子名称: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | 著者 | Wang, X, Xu, C. | | 登録日 | 2022-02-28 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
4E3B
 
 | |
6TT3
 
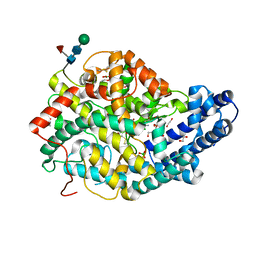 | |
6QLY
 
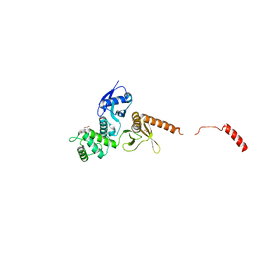 | | IDOL FERM domain | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | 著者 | Martinelli, L, Sixma, T.K. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6TT1
 
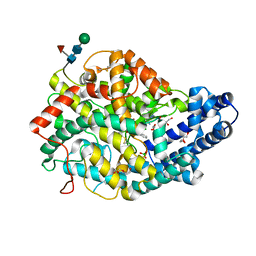 | |
6TT4
 
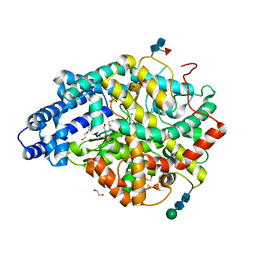 | |
7E83
 
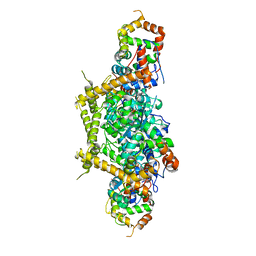 | |
7E84
 
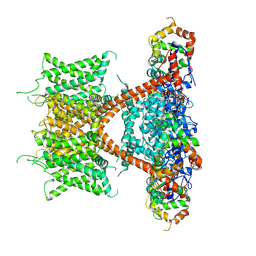 | | CryoEM structure of human Kv4.2-KChIP1 complex | | 分子名称: | Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | 著者 | Kise, Y, Nureki, O. | | 登録日 | 2021-02-28 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8E
 
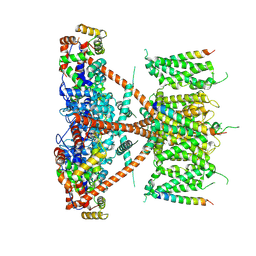 | |
7E8H
 
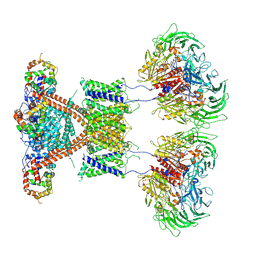 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | 分子名称: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | 著者 | Kise, Y, Nureki, O. | | 登録日 | 2021-03-01 | | 公開日 | 2021-10-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
8CIH
 
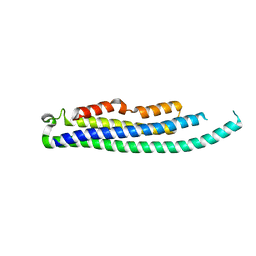 | | Structure of FL CINP | | 分子名称: | Cyclin-dependent kinase 2-interacting protein, SODIUM ION | | 著者 | Foglizzo, M, Zeqiraj, E. | | 登録日 | 2023-02-09 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
7F3F
 
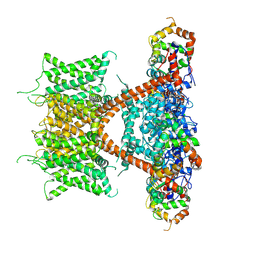 | | CryoEM structure of human Kv4.2-KChIP1 complex | | 分子名称: | Isoform 2 of Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | 著者 | Kise, Y, Nureki, O. | | 登録日 | 2021-06-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
6QLZ
 
 | | IDOL F3ab subdomain | | 分子名称: | E3 ubiquitin-protein ligase MYLIP | | 著者 | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.343 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6HHO
 
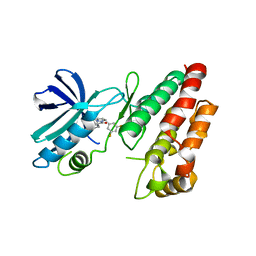 | | Crystal structure of RIP1 kinase in complex with GSK547 | | 分子名称: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | 著者 | Thorpe, J.H, Harris, P.A. | | 登録日 | 2018-08-28 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
2AC5
 
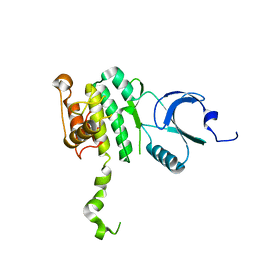 | | Structure of human Mnk2 Kinase Domain mutant D228G | | 分子名称: | MAP kinase-interacting serine/threonine kinase 2, ZINC ION | | 著者 | Jauch, R, Wahl, M.C, Jakel, S, Schreiter, K, Aicher, B, Jackle, H. | | 登録日 | 2005-07-18 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structures of the Mnk2 kinase domain reveal an inhibitory conformation and a zinc binding site.
Structure, 13, 2005
|
|
2AC3
 
 | | Structure of human Mnk2 Kinase Domain | | 分子名称: | MAP kinase-interacting serine/threonine kinase 2, ZINC ION | | 著者 | Jauch, R, Wahl, M.C, Netter, C, Jakel, S, Schreiter, K, Aicher, B, Jackle, H. | | 登録日 | 2005-07-18 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the Mnk2 kinase domain reveal an inhibitory conformation and a zinc binding site.
Structure, 13, 2005
|
|
