7DO6
 
 | |
4HB9
 
 | |
3FOQ
 
 | |
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | 著者 | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | 登録日 | 2005-09-26 | | 公開日 | 2005-10-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
4ZOC
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorotriose | | 分子名称: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
3FSJ
 
 | | Crystal structure of benzoylformate decarboxylase in complex with the inhibitor MBP | | 分子名称: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CALCIUM ION | | 著者 | Brandt, G.S, Kenyon, G.L, McLeish, M.J, Jordan, F, Petsko, G.A, Ringe, D. | | 登録日 | 2009-01-09 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Snapshot of a reaction intermediate: analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor.
Biochemistry, 48, 2009
|
|
5E1Q
 
 | | Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac | | 分子名称: | CALCIUM ION, GLYCEROL, Retaining alpha-galactosidase, ... | | 著者 | Matsunaga, K, Yamashita, K, Tagami, T, Yao, M, Okuyama, M, Kimura, A. | | 登録日 | 2015-09-30 | | 公開日 | 2016-10-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.943 Å) | | 主引用文献 | Efficient synthesis of alpha-galactosyl oligosaccharides using a mutant Bacteroides thetaiotaomicron retaining alpha-galactosidase (BtGH97b).
FEBS J., 284, 2017
|
|
1U1D
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(phenylthio)acyclouridine (ptau) | | 分子名称: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLTHIO)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Bu, W, Settembre, E.C, Ealick, S.E. | | 登録日 | 2004-07-15 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4ZOD
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lin1840 protein, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4L47
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U on the Ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | 登録日 | 2013-06-07 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.220001 Å) | | 主引用文献 | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5QOK
 
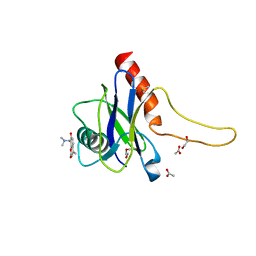 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000294a | | 分子名称: | 1,2-ETHANEDIOL, 8-[(dimethylamino)methyl]-4-methyl-7-oxidanyl-chromen-2-one, ACETATE ION, ... | | 著者 | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | 登録日 | 2019-02-22 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
3CQY
 
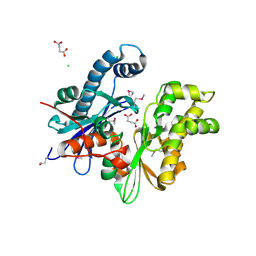 | | Crystal structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1 | | 分子名称: | Anhydro-N-acetylmuramic acid kinase, CHLORIDE ION, SUCCINIC ACID | | 著者 | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-04-03 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1.
To be Published
|
|
2RHM
 
 | |
5TZR
 
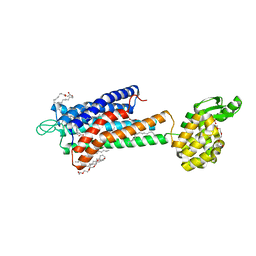 | | GPR40 in complex with partial agonist MK-8666 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | 著者 | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | 登録日 | 2016-11-22 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4LFZ
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U in the Absence of Paromomycin | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | 登録日 | 2013-06-27 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.92000055 Å) | | 主引用文献 | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1T5Q
 
 | | Solution Structure of GIP(1-30)amide in TFE/Water | | 分子名称: | Gastric inhibitory polypeptide | | 著者 | Alana, I, Hewage, C.M, Malthouse, J.P.G, Parker, J.C, Gault, V.A, O'Harte, F.P.M. | | 登録日 | 2004-05-05 | | 公開日 | 2004-11-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the glucose-dependent insulinotropic polypeptide fragment, GIP(1-30)amide.
Biochem.Biophys.Res.Commun., 325, 2004
|
|
1T61
 
 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | 登録日 | 2004-05-05 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
5QOI
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000213a | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | 著者 | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | 登録日 | 2019-02-22 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
1TG4
 
 | | Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution | | 分子名称: | FLAYK peptide, Phospholipase A2, SULFATE ION | | 著者 | Singh, N, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Ethayathulla, A.S, Singh, T.P. | | 登録日 | 2004-05-28 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution
TO BE PUBLISHED
|
|
4H9C
 
 | | Radiation damage study of lysozyme - 0.77 MGy | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | 著者 | Sutton, K.A, Snell, E.H. | | 登録日 | 2012-09-24 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.1998 Å) | | 主引用文献 | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6F25
 
 | | Crystal structure of human acetylcholinesterase in complex with C35. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Dias, J, Nachon, F. | | 登録日 | 2017-11-23 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.05199647 Å) | | 主引用文献 | New evidence for dual binding site inhibitors of acetylcholinesterase as improved drugs for treatment of Alzheimer's disease.
Neuropharmacology, 155, 2019
|
|
3FQL
 
 | | Hepatitis C virus polymerase NS5B (CON1 1-570) with HCV-796 inhibitor | | 分子名称: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, GLYCEROL, RNA-directed RNA polymerase | | 著者 | Harris, S.F, Wong, A. | | 登録日 | 2009-01-07 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus.
J.Biol.Chem., 284, 2009
|
|
3RWQ
 
 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | 分子名称: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | 著者 | Kazmirski, S, Kohls, D. | | 登録日 | 2011-05-09 | | 公開日 | 2011-11-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
4OQG
 
 | | Crystal structure of TEM-1 beta-lactamase in complex with boron-based inhibitor EC25 | | 分子名称: | 3-[(2R)-2-{[(2R)-2-amino-2-phenylacetyl]amino}-2-(dihydroxyboranyl)ethyl]benzoic acid, Ampicillin resistance protein, ZINC ION | | 著者 | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | 登録日 | 2014-02-09 | | 公開日 | 2015-05-20 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
3G2Y
 
 | |
