3L01
 
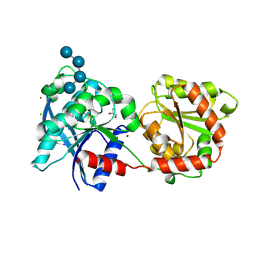 | | Crystal structure of monomeric glycogen synthase from Pyrococcus abyssi | | 分子名称: | CHLORIDE ION, GLYCEROL, GlgA glycogen synthase, ... | | 著者 | Diaz, A, Martinez-Pons, C, Fita, I, Ferrer, J.C, Guinovart, J.J. | | 登録日 | 2009-12-09 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Processivity and Subcellular Localization of Glycogen Synthase Depend on a Non-catalytic High Affinity Glycogen-binding Site.
J.Biol.Chem., 286, 2011
|
|
3ONY
 
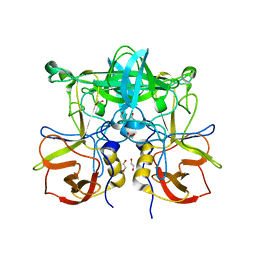 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with Fucose | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose | | 著者 | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | 登録日 | 2010-08-30 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
3L36
 
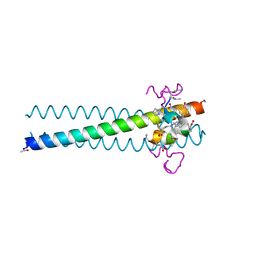 | | PIE12 D-peptide against HIV entry | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | 著者 | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | 登録日 | 2009-12-16 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
1X9I
 
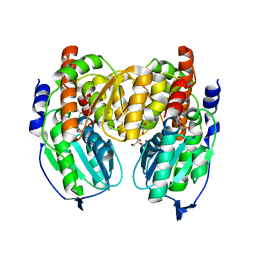 | | Crystal structure of Crystal structure of phosphoglucose/phosphomannose phosphoglucose/phosphomannoseisomerase from Pyrobaculum aerophilum in complex with glucose 6-phosphate | | 分子名称: | GLUCOSE-6-PHOSPHATE, GLYCEROL, glucose-6-phosphate isomerase | | 著者 | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | 登録日 | 2004-08-21 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Structural basis for phosphomannose isomerase activity in phosphoglucose isomerase from Pyrobaculum aerophilum: a subtle difference between distantly related enzymes.
Biochemistry, 43, 2004
|
|
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | 分子名称: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | 著者 | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | 登録日 | 2001-09-25 | | 公開日 | 2001-11-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
1NAR
 
 | |
3ZJ8
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-2 | | 分子名称: | (1R,2S,3S,4R,5R)-4-[(4-bromophenyl)methylamino]-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
2Q7E
 
 | | The structure of pyrrolysyl-tRNA synthetase bound to an ATP analogue | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Kavran, J.M, Steitz, T.A. | | 登録日 | 2007-06-06 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OX9
 
 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | 分子名称: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | 著者 | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | 登録日 | 2014-02-04 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.8035 Å) | | 主引用文献 | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O71
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with FLAVOPIRIDOL | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, Bromodomain-containing protein 4 | | 著者 | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | 登録日 | 2013-12-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
3PL2
 
 | |
2Q66
 
 | |
4O75
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with FOSTAMATINIB | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [6-({5-fluoro-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}amino)-2,2-dimethyl-3-oxo-2,3-dihydro-4H-pyrido[3,2-b][1,4]oxazin-4-yl]methyl dihydrogen phosphate | | 著者 | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | 登録日 | 2013-12-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
2Q7G
 
 | | Pyrrolysine tRNA Synthetase bound to a pyrrolysine analogue (cyc) and ATP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Kavran, J.M, Steitz, T.A. | | 登録日 | 2007-06-06 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1XVX
 
 | | Crystal Structure of iron-loaded Yersinia enterocolitica YfuA | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, FE (III) ION, ... | | 著者 | Shouldice, S.R, McRee, D.E, Dougan, D.R, Tari, L.W, Schryvers, A.B. | | 登録日 | 2004-10-28 | | 公開日 | 2004-12-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Novel Anion-independent Iron Coordination by Members of a Third Class of Bacterial Periplasmic Ferric Ion-binding Proteins
J.Biol.Chem., 280, 2005
|
|
1XIY
 
 | | Crystal Structure of Plasmodium falciparum antioxidant protein (1-Cys peroxiredoxin) | | 分子名称: | peroxiredoxin | | 著者 | Sarma, G.N, Fischer, M, Nickel, C, Becker, K, Karplus, P.A. | | 登録日 | 2004-09-22 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a novel Plasmodium falciparum 1-Cys peroxiredoxin.
J.Mol.Biol., 346, 2005
|
|
7GST
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000056a | | 分子名称: | 1-(methanesulfonyl)-1,2,3,4-tetrahydroquinoline, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Mehlman, T, Ginn, H.M, Keedy, D.A. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSZ
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMSOA000686b | | 分子名称: | 1-[4-methyl-2-(pyridin-4-yl)-1,3-thiazol-5-yl]methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Mehlman, T, Ginn, H.M, Keedy, D.A. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTN
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000625a | | 分子名称: | 1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Mehlman, T, Ginn, H.M, Keedy, D.A. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
2C0L
 
 | |
4BEK
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | 分子名称: | (4S)-4-(4-methoxyphenyl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | 著者 | Banner, D.W, Benz, J, Stihle, M. | | 登録日 | 2013-03-11 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3CLL
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E mutant | | 分子名称: | Aquaporin | | 著者 | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Trnroth-Horsefield, S. | | 登録日 | 2008-03-19 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
1REG
 
 | |
4ONW
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea | | 分子名称: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ACETATE ION, ... | | 著者 | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-01-29 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
