7ZME
 
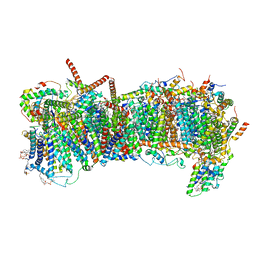 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
6WDD
 
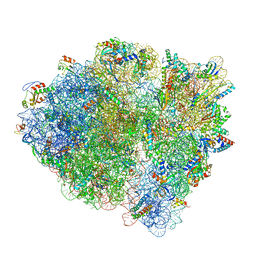 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure V-A) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Demo, G, Korostelev, A.A. | | 登録日 | 2020-03-31 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
7ZMG
 
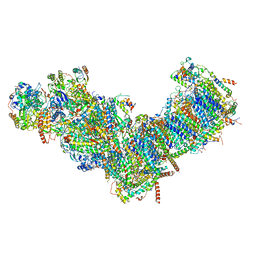 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
8BO0
 
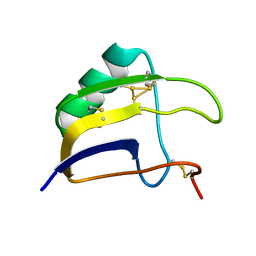 | | Solution structure of Lqq4 toxin from Leiurus quinquestriatus quinquestriatus | | 分子名称: | Alpha-toxin Lqq4 | | 著者 | Mineev, K.S, Motov, V.V, Vassilevski, A.A, Chernykh, M.A, Kuzmenkov, A.I. | | 登録日 | 2022-11-14 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A scorpion toxin affecting sodium channels shows double cis-trans isomerism.
Febs Lett., 597, 2023
|
|
8BU2
 
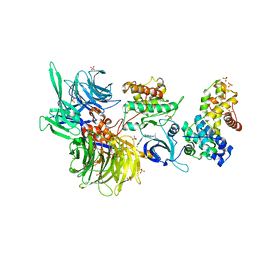 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUF
 
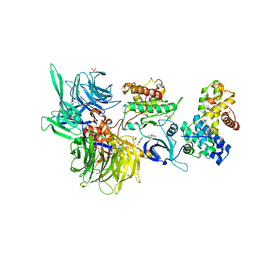 | | Structure of DDB1 bound to Z12-engaged CDK12-cyclin K | | 分子名称: | 2-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylmethyl)-6,7-dimethoxy-3~{H}-quinazolin-4-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | 分子名称: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
8BUP
 
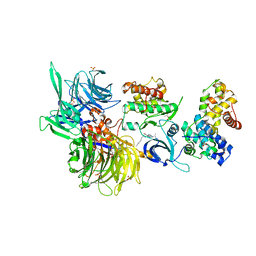 | | Structure of DDB1 bound to DS30-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[3-(3-methylphenyl)propylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7UPL
 
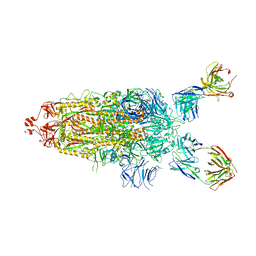 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Patel, A, Ortlund, E. | | 登録日 | 2022-04-15 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
6WIH
 
 | | N-terminal mutation of ISCU2 (L35H36) traps Nfs1 Cys loop in the active site of ISCU2 without metal present. Structure of human mitochondrial complex Nfs1-ISCU2(L35H36)-ISD11 with E.coli ACP1 at 1.9 A resolution (NIAU)2. | | 分子名称: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Acyl carrier protein, ... | | 著者 | Boniecki, M.T, Cygler, M. | | 登録日 | 2020-04-09 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6WPT
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | 著者 | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | 分子名称: | peptide 36-31-3-RD | | 著者 | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | 登録日 | 2020-04-17 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
2GRE
 
 | |
7V20
 
 | |
6WM3
 
 | | Human V-ATPase in state 2 with SidK and ADP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | 著者 | Wang, L, Wu, H, Fu, T.M. | | 登録日 | 2020-04-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
7V23
 
 | |
8BUD
 
 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6WXE
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Upright conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | 著者 | Herrmann, T, Harrison, S.C, Jenni, S. | | 登録日 | 2020-05-10 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
7UDH
 
 | | Integrin alaphIIBbeta3 complex with BMS4-3 | | 分子名称: | (4-{[(5S)-3-(piperidin-4-yl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | 著者 | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | 登録日 | 2022-03-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.99998844 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UFH
 
 | | Integrin alaphIIBbeta3 complex with fradafiban (Mn/Ca) | | 分子名称: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | 登録日 | 2022-03-22 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.99768162 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UK9
 
 | | Integrin alaphIIBbeta3 complex with lamifiban (Mn) | | 分子名称: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | 登録日 | 2022-03-31 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.60001969 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
8BUE
 
 | | Structure of DDB1 bound to Z11-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7UGP
 
 | |
7UKT
 
 | | Integrin alaphIIBbeta3 complex with BMS4.2 | | 分子名称: | (1-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperidin-4-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | 著者 | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | 登録日 | 2022-04-01 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.36994433 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UCY
 
 | | Integrin alaphIIBbeta3 complex with gantofiban | | 分子名称: | (4-{[(5R)-3-(4-carbamimidoylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | 著者 | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | 登録日 | 2022-03-17 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.34996319 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
