1ISW
 
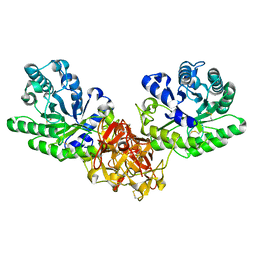 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose | | 分子名称: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
9B0O
 
 | |
1GA3
 
 | |
1ISZ
 
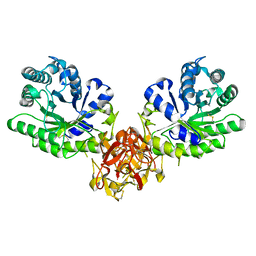 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | 分子名称: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
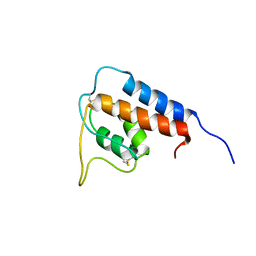
3QQN
 
 | | The retinal specific CD147 Ig0 domain: from molecular structure to biological activity | | 分子名称: | Basigin | | 著者 | Redzic, J.S, Armstrong, G.S, Isern, N.G, Kieft, J.S, Eisenmesser, E.Z, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2011-02-15 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | The Retinal Specific CD147 Ig0 Domain: From Molecular Structure to Biological Activity.
J.Mol.Biol., 411, 2011
|
|
6FJM
 
 | | tubulin-Disorazole Z complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Disorazole Z, ... | | 著者 | Menchon, G, Prota, A.E, Lucena Agell, D, Bucher, P, Jansen, R, Irschik, H, Mueller, R, Paterson, I, Diaz, J.F, Altmann, K.-H, Steinmetz, M.O. | | 登録日 | 2018-01-22 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin.
Nat Commun, 9, 2018
|
|
1IT0
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose | | 分子名称: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISV
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose | | 分子名称: | beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISX
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose | | 分子名称: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISY
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose | | 分子名称: | beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
2D1Z
 
 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D23
 
 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D22
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
7YG6
 
 | | Cryo-EM structure of the EfPiwi(N959K) in complex with piRNA | | 分子名称: | MAGNESIUM ION, Piwi, piRNA | | 著者 | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | 登録日 | 2022-07-11 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFX
 
 | | Cryo-EM structure of Hili in complex with piRNA | | 分子名称: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | 著者 | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | 登録日 | 2022-07-09 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YGN
 
 | | Cryo-EM structure of the Mili in complex with piRNA | | 分子名称: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | 著者 | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | 登録日 | 2022-07-11 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
4GEG
 
 | | Crystal Structure of E.coli MenH Y85F Mutant | | 分子名称: | 1,2-ETHANEDIOL, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, ... | | 著者 | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | 登録日 | 2012-08-01 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
2FBW
 
 | | Avian respiratory complex II with carboxin bound | | 分子名称: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | 著者 | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | 登録日 | 2005-12-10 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
2D24
 
 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2H5X
 
 | | RuvA from Mycobacterium tuberculosis | | 分子名称: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | 著者 | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Alipio, E.Z, Kim, C.Y, Waldo, G.S, Terwilliger, T.C, Segelke, B, Lekin, T, Toppani, D, Hung, L.W, Yu, M, Bursey, E, Muniyappa, K, Chandra, N.R, Vijayan, M. | | 登録日 | 2006-05-28 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Mycobacterium tuberculosis RuvA, a protein involved in recombination.
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2MVZ
 
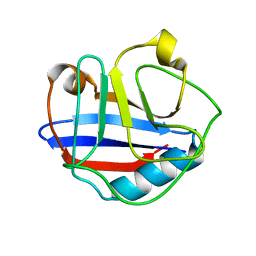 | | Solution Structure for Cyclophilin A from Geobacillus Kaustophilus | | 分子名称: | Peptidyl-prolyl cis-trans isomerase | | 著者 | Holliday, M.J, Isern, N.G, Geoffrey, A.S, Zhang, F, Eisenmesser, E.Z. | | 登録日 | 2014-10-20 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Dynamics of GeoCyp: A Thermophilic Cyclophilin with a Novel Substrate Binding Mechanism That Functions Efficiently at Low Temperatures.
Biochemistry, 54, 2015
|
|
6AQX
 
 | |
6AQV
 
 | |
6AQT
 
 | |
