6W52
 
 | | Prefusion RSV F bound by neutralizing antibody RSB1 | | 分子名称: | Fusion glycoprotein F0, Fusion glycoprotein F1 fused with Fibritin trimerization domain, RSB1 Fab Heavy Chain, ... | | 著者 | Harshbarger, W, Chandramouli, S, Malito, M. | | 登録日 | 2020-03-12 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.74 Å) | | 主引用文献 | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
1ZN8
 
 | | Human Adenine Phosphoribosyltransferase Complexed with AMP, in Space Group P1 at 1.76 A Resolution | | 分子名称: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CHLORIDE ION | | 著者 | Iulek, J, Silva, M, Tomich, C.H.T.P, Thiemann, O.H. | | 登録日 | 2005-05-11 | | 公開日 | 2006-04-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Complexes of Human Adenine Phosphoribosyltransferase Reveal Novel Features of the APRT Catalytic Mechanism
J.Biomol.Struct.Dyn., 25, 2008
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | 分子名称: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | 著者 | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-17 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | 分子名称: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | 分子名称: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
6DDD
 
 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-5 | | 分子名称: | 2,2-dichloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | 著者 | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | 登録日 | 2018-05-10 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6DIM
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 from cocktail soak | | 分子名称: | 1,2-ETHANEDIOL, 4-hydroxyquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | 著者 | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | 登録日 | 2018-05-23 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
7SK7
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, and an extracellular Fab | | 分子名称: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Anti-Fab nanobody, Atypical chemokine receptor 3, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK5
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab | | 分子名称: | Anti-Fab nanobody, Atypical chemokine receptor 3, CHOLESTEROL, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
4RAZ
 
 | |
7JHP
 
 | |
7JN0
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-08-03 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
5BW9
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | 著者 | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | 登録日 | 2015-06-06 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (7 Å) | | 主引用文献 | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
6U81
 
 | |
6U9S
 
 | | Crystal structure of human CD81 large extracellular loop in complex with 5A6 Fab | | 分子名称: | 5A6 FAB Heavy Chain, 5A6 FAB Light Chain, CD81 antigen, ... | | 著者 | Susa, K.J, Seegar, T.C.M, Blacklow, S.C.B, Kruse, A.C. | | 登録日 | 2019-09-09 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking.
Elife, 9, 2020
|
|
6UBS
 
 | | Full length Glycine receptor reconstituted in lipid nanodisc in Apo/Resting conformation | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2019-09-12 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
4JE3
 
 | |
4RE9
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | 著者 | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | 登録日 | 2014-09-22 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.908 Å) | | 主引用文献 | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
7QRW
 
 | |
6YKK
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 15 | | 分子名称: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | 著者 | Pang, L, Strelkov, S.V, Weeks, S.D. | | 登録日 | 2020-04-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.236 Å) | | 主引用文献 | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
6YKV
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 11g | | 分子名称: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | 著者 | Pang, L, Strelkov, S.V, Weeks, S.D. | | 登録日 | 2020-04-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
6DAT
 
 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-02 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.35002637 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6D84
 
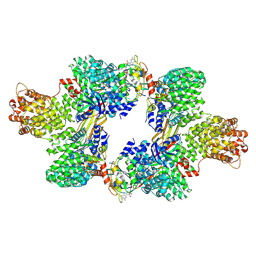 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef (L164A, L165A) dileucine mutant dimer | | 分子名称: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | 著者 | Buffalo, C.Z, Morris, K.L, Hurley, J.H. | | 登録日 | 2018-04-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (6.72 Å) | | 主引用文献 | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
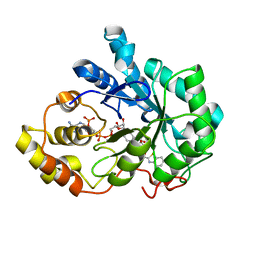
4QR6
 
 | |
