6P3C
 
 | |
6OZX
 
 | | Wild type GapR crystal structure 1 from C. crescentus | | 分子名称: | UPF0335 protein CC_3319 | | 著者 | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | 登録日 | 2019-05-16 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
5FBH
 
 | | Crystal structure of the extracellular domain of human calcium sensing receptor with bound Gd3+ | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | 著者 | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | 登録日 | 2015-12-14 | | 公開日 | 2016-06-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | 著者 | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | 登録日 | 2014-12-16 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.004 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
7SPR
 
 | |
6P0S
 
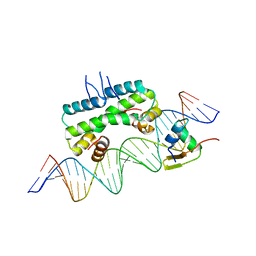 | | Crystal structure of ternary DNA complex "FX2" containing E. coli Fis and phage lambda Xis | | 分子名称: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | 著者 | Hancock, S.P, Cascio, D, Johnson, R.C. | | 登録日 | 2019-05-17 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
6P7T
 
 | |
6YRA
 
 | |
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | 著者 | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | 登録日 | 2020-04-20 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
1BJ3
 
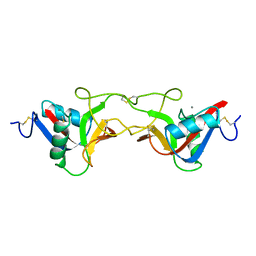 | | CRYSTAL STRUCTURE OF COAGULATION FACTOR IX-BINDING PROTEIN (IX-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | 分子名称: | CALCIUM ION, PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN A), PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN B) | | 著者 | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H, Atoda, H, Morita, T. | | 登録日 | 1998-07-02 | | 公開日 | 1999-08-16 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 A: implication of central loop swapping based on deletion in the linker region.
J.Mol.Biol., 289, 1999
|
|
5W6G
 
 | |
7QHL
 
 | | Crystal structure of Cyclin-dependent kinase 2/cyclin A in complex with 3,5,7-Substituted pyrazolo[4,3-d]pyrimidine inhibitor 24 | | 分子名称: | 1,2-ETHANEDIOL, 5-(2-amino-1-ethyl)thio-3-cyclobutyl-7-[4-(pyrazol-1-yl)benzyl]amino-1(2)H-pyrazolo[4,3-d]pyrimidine, Cyclin-A2, ... | | 著者 | Djukic, S, Skerlova, J, Rezacova, P. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 3,5,7-Substituted Pyrazolo[4,3- d ]Pyrimidine Inhibitors of Cyclin-Dependent Kinases and Cyclin K Degraders.
J.Med.Chem., 65, 2022
|
|
6HVN
 
 | | CdaA-APO Y187A Mutant | | 分子名称: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Heidemann, J.L, Neumann, P, Ficner, R. | | 登録日 | 2018-10-11 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.234 Å) | | 主引用文献 | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
6PIF
 
 | | V. cholerae TniQ-Cascade complex, open conformation | | 分子名称: | Cas7, type I-F CRISPR-associated protein, TniQ monomer 1, ... | | 著者 | Halpin-Healy, T, Klompe, S, Sternberg, S.H. | | 登録日 | 2019-06-26 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Nature, 577, 2020
|
|
6P8C
 
 | | 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (MthRED) from Methanothermobacter thermautotrophicus | | 分子名称: | 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5'-phosphate reductase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Carbone, V, Schofield, L.R, Hannus, I, Sutherland-Smith, A.J, Ronimus, R.S. | | 登録日 | 2019-06-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | The Crystal Structure of 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (MthRED) from Methanothermobacter thermautotrophicus
To Be Published
|
|
4X9G
 
 | | Crystal structure of Dscam1 isoform 6.44, N-terminal four Ig domains | | 分子名称: | Down Syndrome Cell Adhesion Molecule isoform 6.44, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | 登録日 | 2014-12-11 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.403 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
3IW7
 
 | | Human p38 MAP Kinase in Complex with an Imidazo-pyridine | | 分子名称: | 2-({4-[(4-benzylpiperidin-1-yl)carbonyl]benzyl}sulfanyl)-3H-imidazo[4,5-c]pyridine, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Gruetter, C, Simard, J.R, Rauh, D. | | 登録日 | 2009-09-02 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
7TVD
 
 | |
6YUW
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | 分子名称: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Ruza, R.R, Hillier, J, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV2
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | 分子名称: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruza, R.R, Hillier, J, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6HQ8
 
 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 with laminarihexaose bound at a surface site | | 分子名称: | 1,2-ETHANEDIOL, BICINE, Beta-1,3-oligosaccharide phosphorylase, ... | | 著者 | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | 登録日 | 2018-09-24 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
4X9I
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, isoform 9.44, ... | | 著者 | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | 登録日 | 2014-12-11 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.904 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | 分子名称: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | 著者 | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | 登録日 | 2019-06-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6OZY
 
 | | Wild type GapR crystal structure 2 from C. crescentus | | 分子名称: | CADMIUM ION, UPF0335 protein CC_3319 | | 著者 | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | 登録日 | 2019-05-16 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.014 Å) | | 主引用文献 | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
6HVM
 
 | |
