6FEW
 
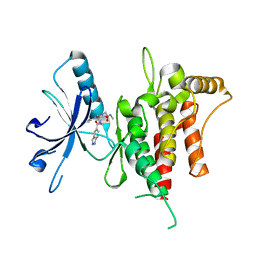 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.440A, P1211, Rfree=24.1% | | 分子名称: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1 | | 著者 | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | 登録日 | 2018-01-03 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIO
 
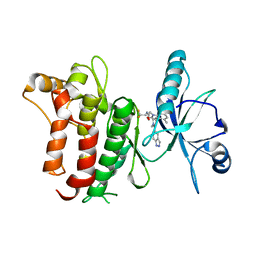 | | DDR1, 2-[1'-(1H-indazole-5-carbonyl)-4-methyl-2-oxospiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.990A, P6522, Rfree=27.7% | | 分子名称: | 2-[1'-(1~{H}-indazol-5-ylcarbonyl)-4-methyl-2-oxidanylidene-spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Epithelial discoidin domain-containing receptor 1 | | 著者 | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | 登録日 | 2018-01-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
2AB1
 
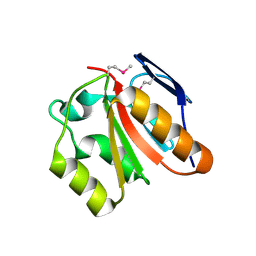 | | X-Ray Structure of Gene Product from Homo Sapiens HS.95870 | | 分子名称: | hypothetical protein | | 著者 | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-07-14 | | 公開日 | 2005-07-26 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | X-Ray Structure of Gene Product from Homo Sapiens HS.95870
To be Published
|
|
5C3I
 
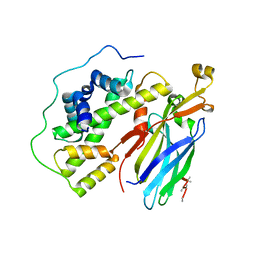 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | 分子名称: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | 著者 | Wang, H, Wang, M, Yang, N, Xu, R.M. | | 登録日 | 2015-06-17 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
5CXB
 
 | |
4FAP
 
 | |
5CXC
 
 | |
5CYK
 
 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | 分子名称: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | 著者 | Wegrecki, M, Bravo, J. | | 登録日 | 2015-07-30 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
6EXQ
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | 分子名称: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | 著者 | Dietzel, U, Kisker, C. | | 登録日 | 2017-11-08 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6F06
 
 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-8-(azetidin-3-yl)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-12,17-dioxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | 分子名称: | (3~{S},14~{E})-8-(azetidin-3-yl)-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, CHLORIDE ION, Cathepsin L1, ... | | 著者 | Kuglstatter, A, Stihle, M. | | 登録日 | 2017-11-17 | | 公開日 | 2018-04-11 | | 最終更新日 | 2018-05-09 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FJO
 
 | | Adenovirus species 26 knob protein, very high resolution | | 分子名称: | Fiber, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | 登録日 | 2018-01-22 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor.
Sci Adv, 5, 2019
|
|
6FKU
 
 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Recombinant protein with shortened C-terminal, in complex with NADP) | | 分子名称: | Aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | 登録日 | 2018-01-24 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
6FJX
 
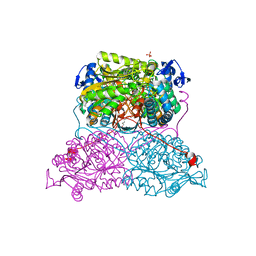 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Native protein) | | 分子名称: | Aldehyde dehydrogenase, SULFATE ION, TRIETHYLENE GLYCOL, ... | | 著者 | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | 登録日 | 2018-01-23 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
6VOU
 
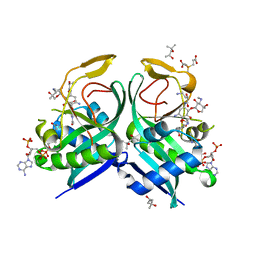 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-plazomicin and CoA | | 分子名称: | (2S)-N-[(1R,2S,3S,4R,5S)-4-{[(2S,3R)-3-(acetylamino)-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-yl]oxy}-5-amino-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-4-amino-2-hydroxybutanamide, (4S)-2-METHYL-2,4-PENTANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, ... | | 著者 | Bassenden, A.V, Berghuis, A.M. | | 登録日 | 2020-01-31 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
6FKV
 
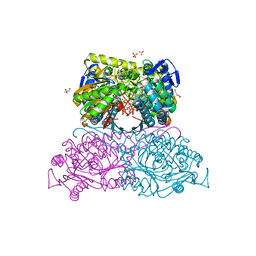 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Recombinant protein with shortened C-terminal, ADH508) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | 登録日 | 2018-01-24 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
4IOX
 
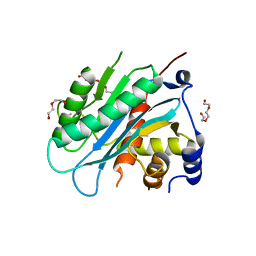 | | The structure of the herpes simplex virus DNA-packaging motor pUL15 C-terminal nuclease domain provides insights into cleavage of concatemeric viral genome precursors | | 分子名称: | ACETATE ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | 著者 | Selvarajan Sigamani, S, Zhao, H, Kamau, Y, Tang, L. | | 登録日 | 2013-01-08 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.458 Å) | | 主引用文献 | The Structure of the Herpes Simplex Virus DNA-Packaging Terminase pUL15 Nuclease Domain Suggests an Evolutionary Lineage among Eukaryotic and Prokaryotic Viruses.
J.Virol., 87, 2013
|
|
4YUU
 
 | | Crystal structure of oxygen-evolving photosystem II from a red alga | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ago, H, Shen, J.-R. | | 登録日 | 2015-03-19 | | 公開日 | 2016-01-20 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (2.7700038 Å) | | 主引用文献 | Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
J.Biol.Chem., 291, 2016
|
|
4UE4
 
 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XEJ
 
 | | IRES bound to bacterial Ribosome | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhu, J, Korostelev, A, Noller, H.F, Donohue, J.P. | | 登録日 | 2014-12-23 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Initiation of translation in bacteria by a structured eukaryotic IRES RNA.
Nature, 519, 2015
|
|
4UE5
 
 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5J25
 
 | |
7TL7
 
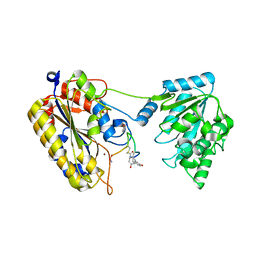 | | 1.90A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Sa-D2) | | 分子名称: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, IMIDAZOLE, SODIUM ION, ... | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | 登録日 | 2022-01-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
7TL8
 
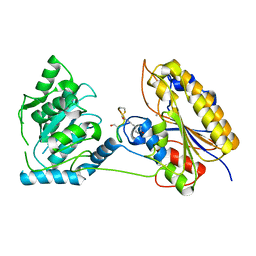 | | 1.95A resolution structure of independent phosphoglycerate mutase from S. aureus in complex with a macrocyclic peptide inhibitor (Sa-D3) | | 分子名称: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION, Peptide Sa-D3 | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | 登録日 | 2022-01-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
7KE2
 
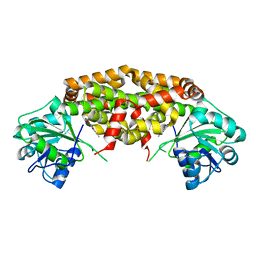 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+ and NSC116565 | | 分子名称: | 6-hydroxy-2-methyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | 著者 | Kurz, J.L, Patel, K.P, Guddat, L.W. | | 登録日 | 2020-10-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Discovery of a Pyrimidinedione Derivative with Potent Inhibitory Activity against Mycobacterium tuberculosis Ketol-Acid Reductoisomerase.
Chemistry, 27, 2021
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | 分子名称: | rRNA expansion segment ES27L in an "out" conformation | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-13 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
