7QGS
 
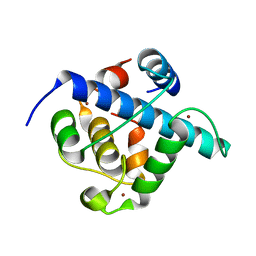 | | Crystal structure of p300 CH1 domain in complex with CITIF (a CITED2-HIF-1alpha hybrid) | | 分子名称: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone acetyltransferase, ZINC ION | | 著者 | Hegedus, Z, Wilson, A.J, Edwards, T.A. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Understanding p300-transcription factor interactions using sequence variation and hybridization.
Rsc Chem Biol, 3, 2022
|
|
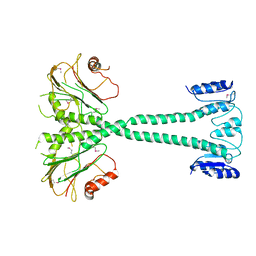
3F7A
 
 | |
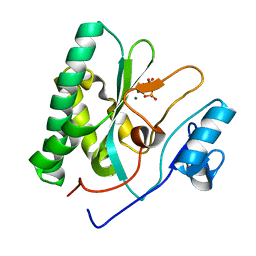
8XIG
 
 | |
7QSM
 
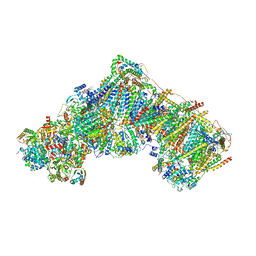 | | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSL
 
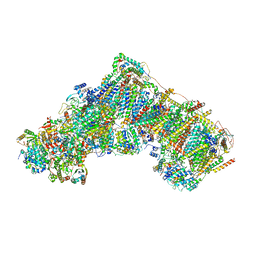 | | Bovine complex I in lipid nanodisc, Active-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSN
 
 | | Bovine complex I in lipid nanodisc, Deactive-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
8XJ6
 
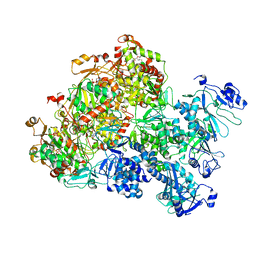 | | The Cryo-EM structure of MPXV E5 apo conformation | | 分子名称: | AMP PHOSPHORAMIDATE, Monkeypox virus E5, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Zhang, W, Liu, Y, Gao, H, Gan, J. | | 登録日 | 2023-12-20 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
7QSK
 
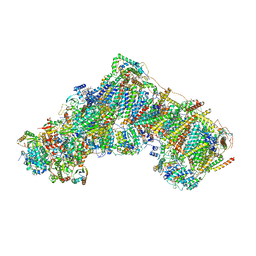 | | Bovine complex I in lipid nanodisc, Active-Q10 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSO
 
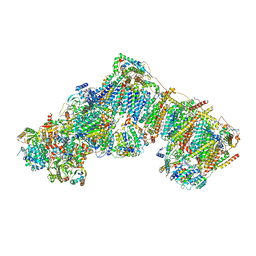 | | Bovine complex I in lipid nanodisc, State 3 (Slack) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7RCB
 
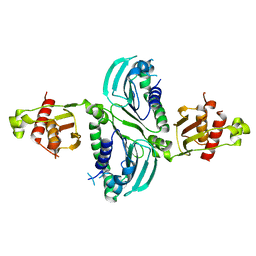 | | Crystal Structure of a PMS2 VUS | | 分子名称: | Mismatch repair endonuclease PMS2 | | 著者 | D'Arcy, B.M, Prakash, A. | | 登録日 | 2021-07-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7RCI
 
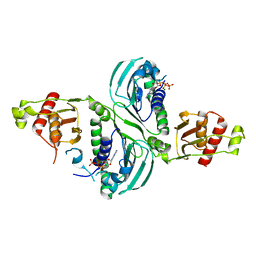 | | Crystal Structure of a PMS2 VUS with Substrate | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | 著者 | D'Arcy, B.M, Prakash, A. | | 登録日 | 2021-07-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7RCK
 
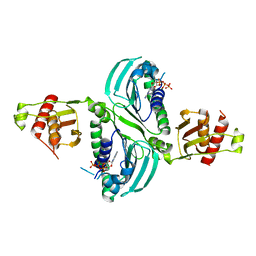 | | Crystal Structure of PMS2 with Substrate | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | 著者 | D'Arcy, B.M, Prakash, A. | | 登録日 | 2021-07-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7SYJ
 
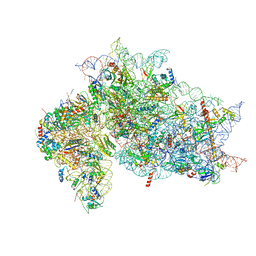 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYX
 
 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYW
 
 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYU
 
 | | Structure of the delta dII IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7QDA
 
 | | Crystal structure of CalpL | | 分子名称: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Schneberger, N, Hagelueken, G. | | 登録日 | 2021-11-26 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-02-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
7SYT
 
 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYH
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
