4TZ4
 
 | | Crystal Structure of Human Cereblon in Complex with DDB1 and Lenalidomide | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, S-Lenalidomide, ... | | 著者 | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B, Riley, M. | | 登録日 | 2014-07-09 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
To Be Published
|
|
8WQR
 
 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | 分子名称: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | 著者 | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | 登録日 | 2023-10-12 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
8BUN
 
 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU4
 
 | | Structure of DDB1 bound to DS22-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6DSZ
 
 | |
6ZUE
 
 | |
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | 分子名称: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | 登録日 | 2024-01-12 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8U17
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Clifton, M.C, Ma, X, Ornelas, E. | | 登録日 | 2023-08-30 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8D7U
 
 | |
8B3F
 
 | | Pol II-CSB-CSA-DDB1-ELOF1 | | 分子名称: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2022-09-16 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Pol II-CSB-CSA-DDB1-ELOF1 structure.
To Be Published
|
|
8D7W
 
 | |
8D7Z
 
 | |
2HYE
 
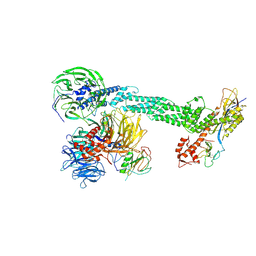 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | 分子名称: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | 著者 | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | 登録日 | 2006-08-05 | | 公開日 | 2006-10-03 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
4A09
 
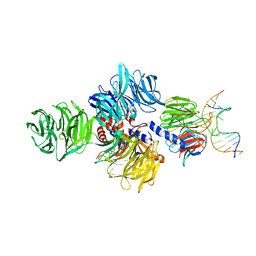 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
8BU2
 
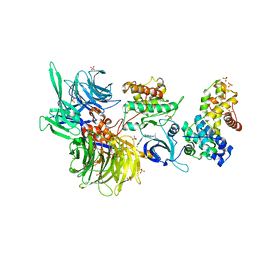 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU5
 
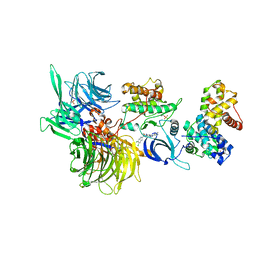 | | Structure of DDB1 bound to SR-4835-engaged CDK12-cyclin K | | 分子名称: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.134 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7U8F
 
 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | 分子名称: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | 著者 | Ma, X, Ornelas, E, Clifton, M.C. | | 登録日 | 2022-03-08 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
8TZX
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | 分子名称: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | 著者 | Clifton, M.C, Ma, X, Ornelas, E. | | 登録日 | 2023-08-28 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Science, 385, 2024
|
|
8T9A
 
 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | 著者 | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | 登録日 | 2023-06-23 | | 公開日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
To Be Published
|
|
8BUA
 
 | | Structure of DDB1 bound to 919278-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-~{N}-(1~{H}-benzimidazol-2-yl)-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.193 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUD
 
 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUQ
 
 | | Structure of DDB1 bound to DS43-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[1-(3-chlorophenyl)pyrazol-3-yl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
5V3O
 
 | | Cereblon in complex with DDB1 and CC-220 | | 分子名称: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Matyskiela, M, Pagarigan, B, Chamberlain, P. | | 登録日 | 2017-03-07 | | 公開日 | 2017-05-03 | | 最終更新日 | 2018-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos.
J. Med. Chem., 61, 2018
|
|
8D7V
 
 | |
4E5Z
 
 | |
