4IO4
 
 | | Crystal Structure of the AvGluR1 ligand binding domain complex with serine at 1.94 Angstrom resolution | | 分子名称: | AvGluR1 ligand binding domain, CHLORIDE ION, GLYCEROL, ... | | 著者 | Lomash, S, Chittori, S, Mayer, M.L. | | 登録日 | 2013-01-07 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Anions Mediate Ligand Binding in Adineta vaga Glutamate Receptor Ion Channels.
Structure, 21, 2013
|
|
2XXU
 
 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with glutamate | | 分子名称: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2XXX
 
 | |
4IO7
 
 | |
2XXR
 
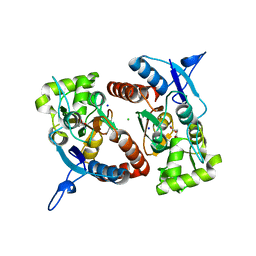 | | Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with glutamate | | 分子名称: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2010-11-11 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
4IO2
 
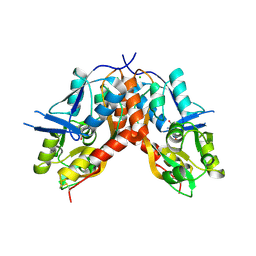 | |
2XXT
 
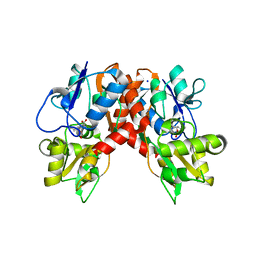 | | Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with kainate | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2XXV
 
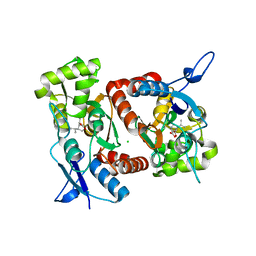 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with kainate | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2XXW
 
 | | Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate | | 分子名称: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
4IY6
 
 | | Crystal structure of the GLUA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and ME-CX516 at 1.72 A resolution | | 分子名称: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | 著者 | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-01-28 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4L17
 
 | | GluA2-L483Y-A665C ligand-binding domain in complex with the antagonist DNQX | | 分子名称: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | 著者 | Lau, A.Y, Blachowicz, L, Roux, B. | | 登録日 | 2013-06-02 | | 公開日 | 2013-08-14 | | 最終更新日 | 2017-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A conformational intermediate in glutamate receptor activation.
Neuron, 79, 2013
|
|
6UCB
 
 | | GluA2 in complex with its auxiliary subunit CNIH3 - with antagonist ZK200775, LBD, TMD, CNIH3, and lipids | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Glutamate receptor 2, ... | | 著者 | Nakagawa, T. | | 登録日 | 2019-09-15 | | 公開日 | 2019-12-04 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structures of the AMPA receptor in complex with its auxiliary subunit cornichon.
Science, 366, 2019
|
|
6UD8
 
 | |
6U6I
 
 | |
6UD4
 
 | |
6USV
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | 分子名称: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | 著者 | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6USU
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with L689,560 and glutamate | | 分子名称: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | 著者 | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, H. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6UZ6
 
 | |
6UZG
 
 | |
6UZX
 
 | | Crystal structure of GLUN1/GLUN2A-4M mutant ligand-binding domain in complex with glycine and UBP791 | | 分子名称: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCEROL, GLYCINE, ... | | 著者 | Wang, J.X, Furukawa, H. | | 登録日 | 2019-11-15 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
6UZR
 
 | |
6VE8
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | 登録日 | 2019-12-30 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VEA
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | 分子名称: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | 著者 | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | 登録日 | 2019-12-30 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6UZW
 
 | | Crystal structure of GLUN1/GLUN2A ligand-binding domain in complex with glycine and UBP791 | | 分子名称: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | 著者 | Wang, J.X, Furukawa, H. | | 登録日 | 2019-11-15 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
6WHS
 
 | | GluN1b-GluN2B NMDA receptor in non-active 1 conformation at 3.95 angstrom resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Chou, T, Tajima, N, Furukawa, H. | | 登録日 | 2020-04-08 | | 公開日 | 2020-08-05 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
