8A8V
 
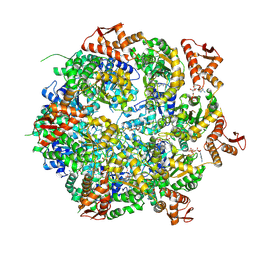 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8U
 
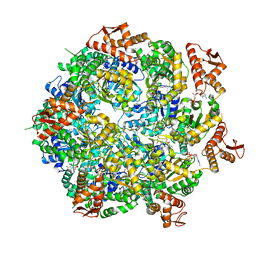 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8W
 
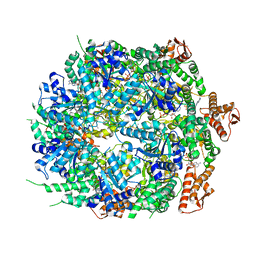 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8AMZ
 
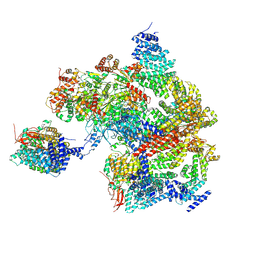 | | Spinach 19S proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1 homolog, 26S proteasome non-ATPase regulatory subunit 2 homolog, 26S proteasome regulatory subunit 7, ... | | 著者 | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | 登録日 | 2022-08-04 | | 公開日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of the plant 26S proteasome
Plant Communications, 3, 2022
|
|
8B5R
 
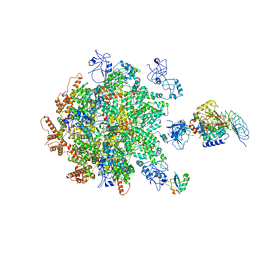 | | p97-p37-SPI substrate complex | | 分子名称: | I3 sequence being threaded through the p97 channel, Protein phosphatase 1 regulatory subunit 7, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit, ... | | 著者 | van den Boom, J, Marini, G, Meyer, H, Saibil, H. | | 登録日 | 2022-09-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structural basis of ubiquitin-independent PP1 complex disassembly by p97.
Embo J., 42, 2023
|
|
8C0V
 
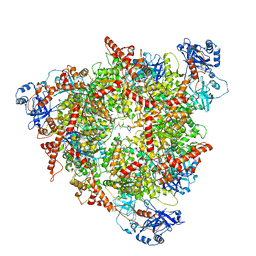 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | 登録日 | 2022-12-19 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0W
 
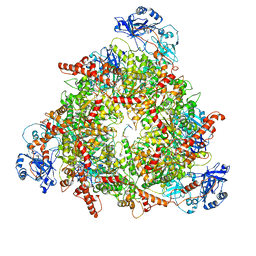 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | 登録日 | 2022-12-19 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8CVT
 
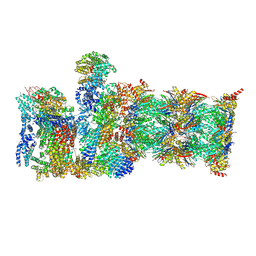 | | Human 19S-20S proteasome, state SD2 | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | 著者 | Zhao, J. | | 登録日 | 2022-05-18 | | 公開日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6X
 
 | |
8D6Y
 
 | |
8DAR
 
 | |
8DAU
 
 | |
8DAW
 
 | |
8DAS
 
 | |
8DAT
 
 | |
8DAV
 
 | |
8DR3
 
 | |
8DR4
 
 | |
8DR7
 
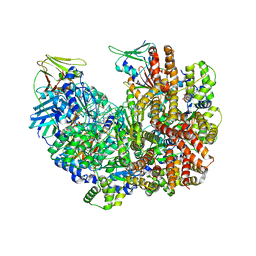 | | Open state of RFC:PCNA bound to a nicked dsDNA | | 分子名称: | DNA (26-MER), DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR5
 
 | |
8DQW
 
 | |
8DR1
 
 | |
8DQX
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR0
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | 分子名称: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQZ
 
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | 分子名称: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
