3J99
 
 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) | | 分子名称: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
8FS4
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
3J3T
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | 分子名称: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS5
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS8
 
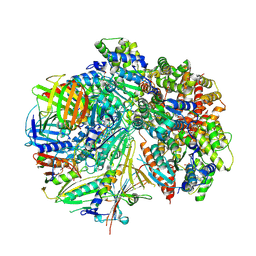 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
3JCO
 
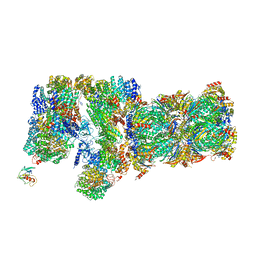 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3J96
 
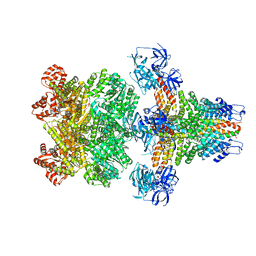 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) | | 分子名称: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | 分子名称: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
8A8V
 
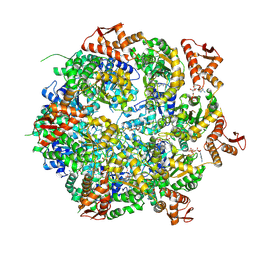 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8U
 
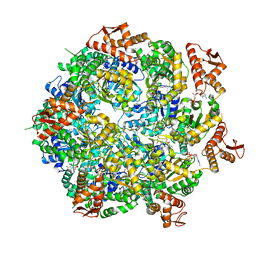 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8GLV
 
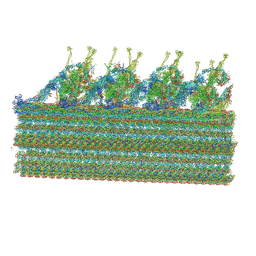 | |
8A8W
 
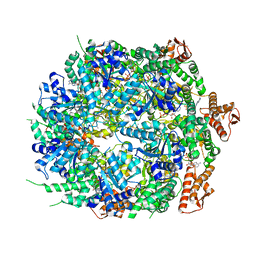 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8B5R
 
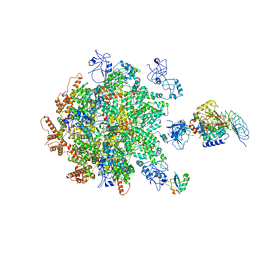 | | p97-p37-SPI substrate complex | | 分子名称: | I3 sequence being threaded through the p97 channel, Protein phosphatase 1 regulatory subunit 7, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit, ... | | 著者 | van den Boom, J, Marini, G, Meyer, H, Saibil, H. | | 登録日 | 2022-09-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structural basis of ubiquitin-independent PP1 complex disassembly by p97.
Embo J., 42, 2023
|
|
8HL7
 
 | |
3J95
 
 | | Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Vesicle-fusing ATPase | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J3U
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | 分子名称: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J94
 
 | | Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vesicle-fusing ATPase | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3JCP
 
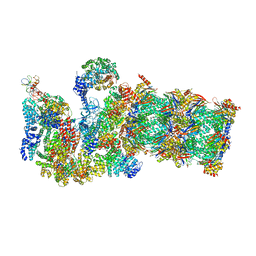 | | Structure of yeast 26S proteasome in M2 state derived from Titan dataset | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1KYI
 
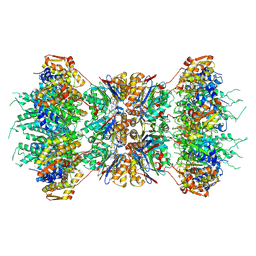 | | HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex | | 分子名称: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ... | | 著者 | Sousa, M.C, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2002-02-04 | | 公開日 | 2002-05-15 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of HslUV Complexed with a Vinyl Sulfone Inhibitor:
Corroboration of a Proposed Mechanism of Allosteric Activation of
HslV by HslU
J.Mol.Biol., 318, 2002
|
|
3HU1
 
 | | Structure of p97 N-D1 R95G mutant in complex with ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Tang, W.-K. | | 登録日 | 2009-06-12 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3HU3
 
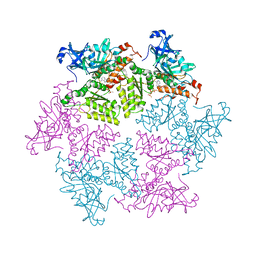 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Tang, W.-K. | | 登録日 | 2009-06-12 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | 分子名称: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J98
 
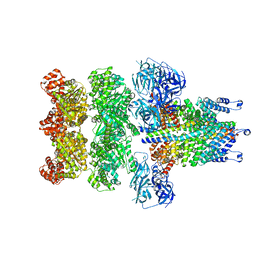 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) | | 分子名称: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
