3E9P
 
 | |
5QZQ
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 41 | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-02-12 | | 公開日 | 2020-06-10 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QZV
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 46 | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-02-12 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7B9V
 
 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | 分子名称: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | 著者 | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | 登録日 | 2020-12-14 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
5DCA
 
 | | Crystal structure of yeast full length Brr2 in complex with Prp8 Jab1 domain | | 分子名称: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2 | | 著者 | Absmeier, E, Wollenhaupt, J, Santos, K.F, Wahl, M.C. | | 登録日 | 2015-08-23 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation.
Genes Dev., 29, 2015
|
|
4BGD
 
 | | Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Nguyen, T.H.D, Li, J, Nagai, K. | | 登録日 | 2013-03-25 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of Brr2-Prp8 Interactions and Implications for U5 Snrnp Biogenesis and the Spliceosome Active Site
Structure, 21, 2013
|
|
6TEO
 
 | | Crystal structure of a yeast Snu114-Prp8 complex | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | 著者 | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | 登録日 | 2019-11-12 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
3ZEF
 
 | | Crystal structure of Prp8:Aar2 complex: second crystal form at 3.1 Angstrom resolution | | 分子名称: | A1 CISTRON-SPLICING FACTOR AAR2, PRE-MRNA-SPLICING FACTOR 8 | | 著者 | Galej, W.P, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2012-12-05 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of Prp8 Reveals Active Site Cavity of the Spliceosome
Nature, 493, 2013
|
|
6J6G
 
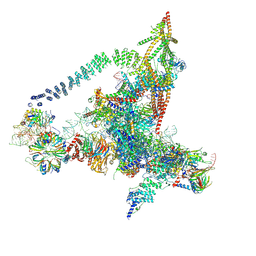 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
3SBG
 
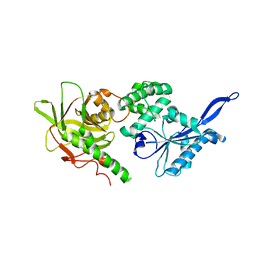 | |
6BK8
 
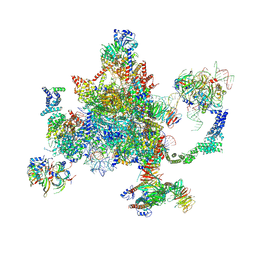 | | S. cerevisiae spliceosomal post-catalytic P complex | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | 著者 | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | 登録日 | 2017-11-07 | | 公開日 | 2018-02-21 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
5GMK
 
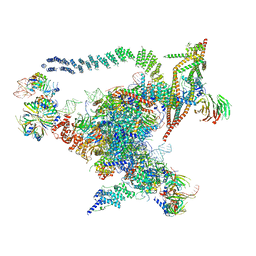 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | 分子名称: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-07-14 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
5M52
 
 | | Crystal structure of yeast Brr2 full-lenght in complex with Prp8 Jab1 domain | | 分子名称: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2 | | 著者 | Wollenhaupt, J, Absmeier, E, Becke, C, Santos, K.F, Wahl, M.C. | | 登録日 | 2016-10-20 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2.
Cell Cycle, 16, 2017
|
|
5ZWM
 
 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.4~4.6 angstrom (tri-snRNP and U2 snRNP Part) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, ... | | 著者 | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2018-05-16 | | 公開日 | 2018-08-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5Y88
 
 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-08-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5GM6
 
 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-07-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
5YLZ
 
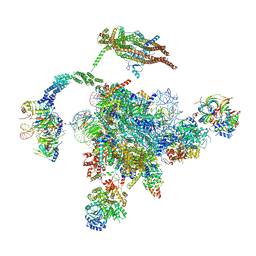 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-10-20 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5GAP
 
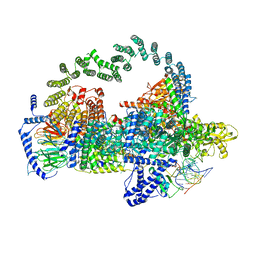 | | Body region of the U4/U6.U5 tri-snRNP | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | 著者 | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | 登録日 | 2015-12-15 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5GAM
 
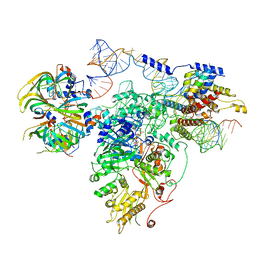 | | Foot region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-splicing factor 8, Pre-mRNA-splicing factor SNU114, ... | | 著者 | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | 登録日 | 2015-12-15 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Angstrom resolution
Nature, 530, 2016
|
|
6J6Q
 
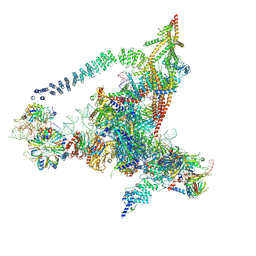 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5GAN
 
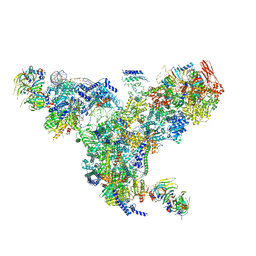 | | The overall structure of the yeast spliceosomal U4/U6.U5 tri-snRNP at 3.7 Angstrom | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-processing factor 31, ... | | 著者 | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | 登録日 | 2015-12-15 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
6EXN
 
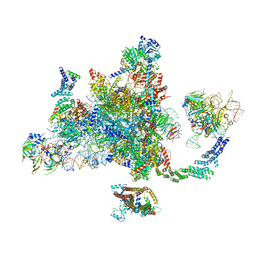 | | Post-catalytic P complex spliceosome with 3' splice site docked | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | 著者 | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | 登録日 | 2017-11-08 | | 公開日 | 2018-01-17 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
5LJ3
 
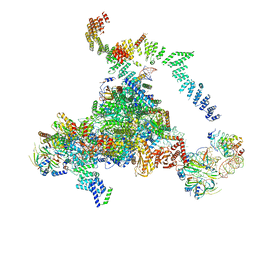 | | Structure of the core of the yeast spliceosome immediately after branching | | 分子名称: | CEF1, CLF1, CWC15, ... | | 著者 | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-03 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
3JCM
 
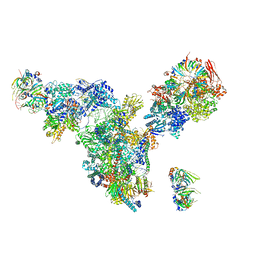 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | 著者 | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | 登録日 | 2015-12-23 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
