1RT3
 
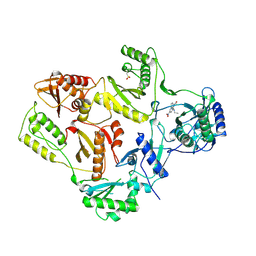 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | 分子名称: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Stammers, D.K, Stuart, D.I. | | 登録日 | 1998-06-29 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1RT7
 
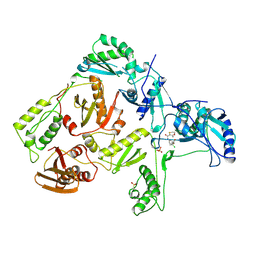 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC84 | | 分子名称: | 1-METHYL ETHYL 1-CHLORO-5-[[(5,6DIHYDRO-2-METHYL-1,4-OXATHIIN-3-YL)CARBONYL]AMINO]BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | 著者 | Ren, J, Stammers, D.K, Stuart, D.I. | | 登録日 | 1998-07-29 | | 公開日 | 1999-07-29 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1T05
 
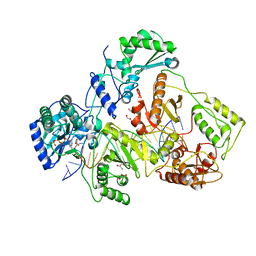 | | HIV-1 reverse transcriptase crosslinked to template-primer with tenofovir-diphosphate bound as the incoming nucleotide substrate | | 分子名称: | GLYCEROL, MAGNESIUM ION, POL polyprotein, ... | | 著者 | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | 登録日 | 2004-04-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
1E6J
 
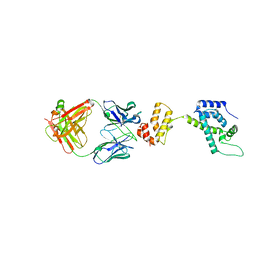 | | Crystal structure of HIV-1 capsid protein (p24) in complex with Fab13B5 | | 分子名称: | CAPSID PROTEIN P24, IMMUNOGLOBULIN | | 著者 | Berthet-Colominas, C, Monaco, S, Novelli, A, Sibai, G, Mallet, F, Cusack, S. | | 登録日 | 2000-08-18 | | 公開日 | 2000-11-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
1JLC
 
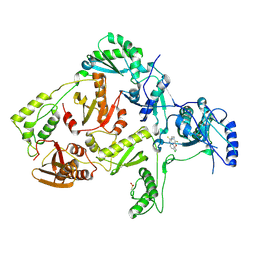 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | 分子名称: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1RTI
 
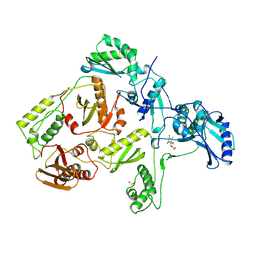 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | 分子名称: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | 登録日 | 1995-05-03 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
6CQR
 
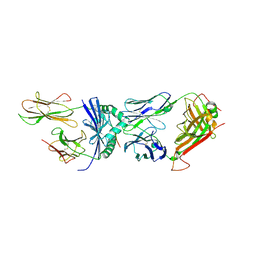 | | Crystal structure of F24 TCR -DR1-RQ13 peptide complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F24 alpha chain, F24 beta chain, ... | | 著者 | Farenc, C, Gras, S, Rossjohn, J. | | 登録日 | 2018-03-16 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
3DM2
 
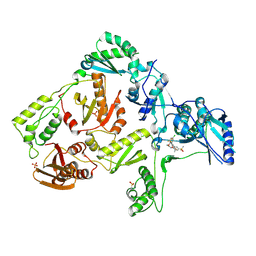 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | 分子名称: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Ren, J, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2008-06-30 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3KJV
 
 | | HIV-1 reverse transcriptase in complex with DNA | | 分子名称: | 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', 5'-D(*AP*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2009-11-03 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
3DRS
 
 | | HIV reverse transcriptase K103N mutant in complex with inhibitor R8D | | 分子名称: | 3-chloro-5-[2-chloro-5-(1H-pyrazolo[3,4-b]pyridin-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase/ribonuclease H, p66 RT | | 著者 | Yan, Y, Prasad, S. | | 登録日 | 2008-07-11 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
2WOM
 
 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (K103N). | | 分子名称: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | 登録日 | 2009-07-27 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1.
Antimicrob. Agents Chemother., 54, 2010
|
|
1RTD
 
 | | STRUCTURE OF A CATALYTIC COMPLEX OF HIV-1 REVERSE TRANSCRIPTASE: IMPLICATIONS FOR NUCLEOSIDE ANALOG DRUG RESISTANCE | | 分子名称: | DNA PRIMER FOR REVERSE TRANSCRIPTASE, DNA TEMPLATE FOR REVERSE TRANSCRIPTASE, MAGNESIUM ION, ... | | 著者 | Chopra, R, Huang, H, Verdine, G.L, Harrison, S.C. | | 登録日 | 1998-08-26 | | 公開日 | 1998-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance.
Science, 282, 1998
|
|
6BSI
 
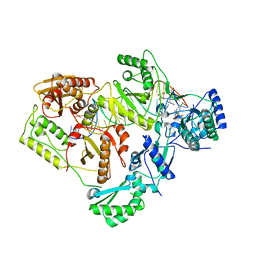 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid containing the polypurine-tract sequence | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*TP*TP*TP*TP*CP*TP*TP*TP*TP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4B3O
 
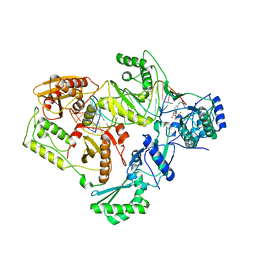 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | 著者 | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | 登録日 | 2012-07-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
6BJ2
 
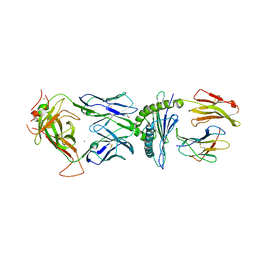 | | TCR589 in complex with HIV(Pol448-456)/HLA-B35 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | 著者 | Jude, K.M, Sibener, L.V, Garcia, K.C. | | 登録日 | 2017-11-03 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | 分子名称: | HIV-1 CA | | 著者 | Summers, B.J, Xiong, Y. | | 登録日 | 2018-08-08 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EC2
 
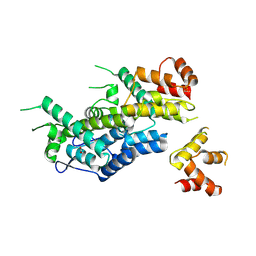 | | Structure of HIV-1 CA 1/3-hexamer | | 分子名称: | ACETATE ION, Capsid protein p24 | | 著者 | Summers, B.J, Xiong, Y. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6SKK
 
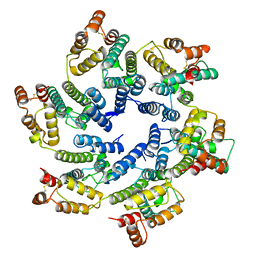 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | capsid protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WAZ
 
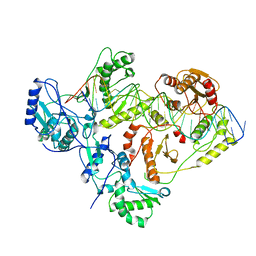 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | 分子名称: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ECO
 
 | |
6WB0
 
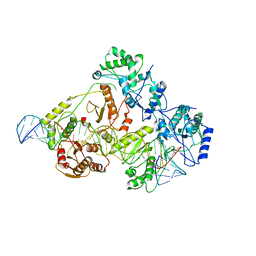 | | +3 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | 分子名称: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
7ASH
 
 | |
6SLQ
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-09 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
