3PHM
 
 | |
3BWE
 
 | | Crystal structure of aggregated form of DJ1 | | Descriptor: | PHOSPHATE ION, Protein DJ-1 | | Authors: | Cha, S.S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
J.Biol.Chem., 283, 2008
|
|
3C75
 
 | | Paracoccus versutus methylamine dehydrogenase in complex with amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, Methylamine dehydrogenase heavy chain, ... | | Authors: | Cavalieri, C, Biermann, N, Vlasie, M.D, Einsle, O, Merli, A, Ferrari, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of crystal and solution states of the 138 kDa complex of methylamine dehydrogenase and amicyanin from Paracoccus versutus.
Biochemistry, 47, 2008
|
|
3ATO
 
 | | Glycine ethyl ester shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Kumasaka, T. | | Deposit date: | 2011-01-06 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Glycine ethyl ester shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation
To be Published
|
|
5OPA
 
 | | The crystal structure of P450 CYP121 in complex with lead compound 7b | | Descriptor: | 3-(4-fluorophenyl)-4-(imidazol-1-ylmethyl)-1-phenyl-pyrazole, DI(HYDROXYETHYL)ETHER, Mycocyclosin synthase, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
6C0T
 
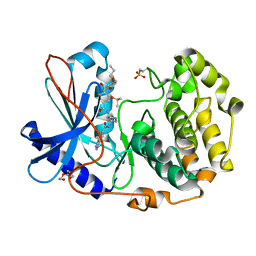 | | Crystal structure of cGMP-dependent protein kinase Ialpha (PKG Ialpha) catalytic domain bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, TRIETHYLENE GLYCOL, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
3Q45
 
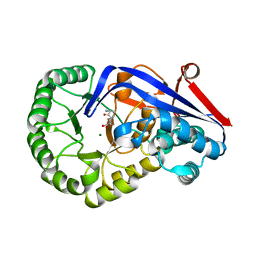 | | Crystal structure of Dipeptide Epimerase from Cytophaga hutchinsonii complexed with Mg and dipeptide D-Ala-L-Val | | Descriptor: | D-ALANINE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family; possible chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2010-12-22 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3PNK
 
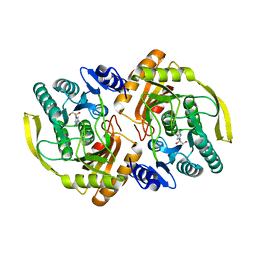 | | Crystal Structure of E.coli Dha kinase DhaK | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6T4C
 
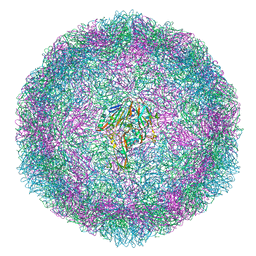 | | Bovine enterovirus F3 in complex with glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
3POC
 
 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
6BYL
 
 | | Structure of 14-3-3 gamma bound to O-GlcNAcylated thr peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSASTTVPVTTATTTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
6ND1
 
 | | CryoEM structure of the Sec Complex from yeast | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Wu, X, Cabanos, C, Rapoport, T.A. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the post-translational protein translocation machinery of the ER membrane.
Nature, 566, 2019
|
|
3PRJ
 
 | |
3PLR
 
 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PTZ
 
 | |
3PNQ
 
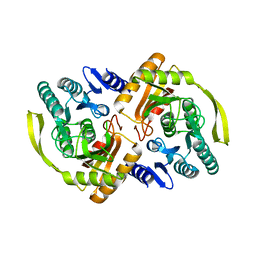 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) complex with Dha | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3APG
 
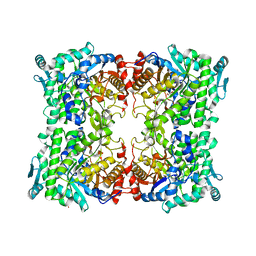 | |
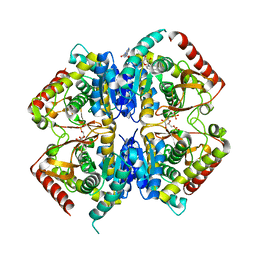
3PQD
 
 | |
6NJD
 
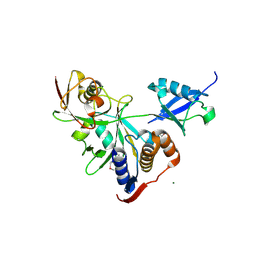 | |
6R76
 
 | |
6BYJ
 
 | | Structure of human 14-3-3 gamma bound to O-GlcNAc peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSTTATPPVSQASSTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
6LXO
 
 | | TvCyP2 in apo form 1 | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
6LXR
 
 | | TvCyP2 in apo form 4 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
1MPU
 
 | | Crystal Structure of the free human NKG2D immunoreceptor | | Descriptor: | NKG2-D type II integral membrane protein, PHOSPHATE ION | | Authors: | McFarland, B.J, Kortemme, T, Baker, D, Strong, R.K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Symmetry Recognizing Asymmetry: Analysis of the Interactions between the C-Type Lectin-like Immunoreceptor NKG2D and MHC
Class I-like Ligands
Structure, 11, 2003
|
|
3ATQ
 
 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, TETRADECANE | | Authors: | Sasaki, D, Fujihashi, M, Murakami, M, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
