6CC0
 
 | |
2NVL
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfonic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4WR8
 
 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-180) | | Descriptor: | 4-[4-(quinolin-2-yl)-1H-1,2,3-triazol-1-yl]phenol, Macrophage migration inhibitory factor, SODIUM ION, ... | | Authors: | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
6CCN
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-2,4-dihydroxy-N-(2-(4-hydroxy-1H-benzo[d]imidazol-2-yl)ethyl)-3,3-dimethylbutanamide | | Descriptor: | (2R)-2,4-dihydroxy-N-[2-(7-hydroxy-1H-benzimidazol-2-yl)ethyl]-3,3-dimethylbutanamide, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
4X0O
 
 | | Beta-ketoacyl-(acyl carrier protein) synthase III-2 (FabH2) from Vibrio cholerae soaked with Acetyl-CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Chordia, M.D, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
5VQR
 
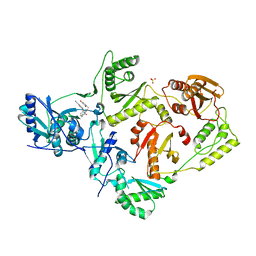 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
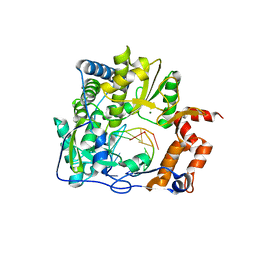
4X2B
 
 | |
2NZI
 
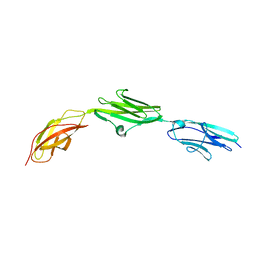 | |
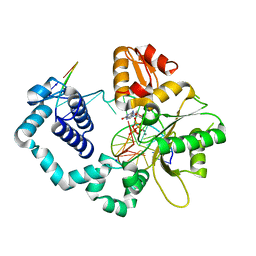
6CUA
 
 | |
6CV6
 
 | |
4X3R
 
 | | Avi-GCPII structure in complex with FITC-conjugated GCPII-specific inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Konvalinka, J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design of highly potent urea-based, exosite-binding inhibitors selective for glutamate carboxypeptidase II.
J.Med.Chem., 58, 2015
|
|
4X4F
 
 | |
4WSA
 
 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
4X4M
 
 | | Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high affinity IgG binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2014-12-03 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.485 Å) | | Cite: | Structure of Fc gamma RI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2NUY
 
 | |
2O24
 
 | |
5VF3
 
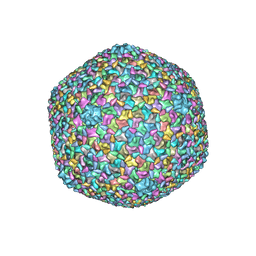 | | Bacteriophage T4 isometric capsid | | Descriptor: | Capsid vertex protein gp24, Highly immunogenic outer capsid protein, Major capsid protein, ... | | Authors: | Chen, Z, Sun, L, Zhang, Z, Fokine, A, Padilla-Sanchez, V, Hanein, D, Jiang, W, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 isometric head at 3.3- angstrom resolution and its relevance to the assembly of icosahedral viruses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2O29
 
 | |
5VV3
 
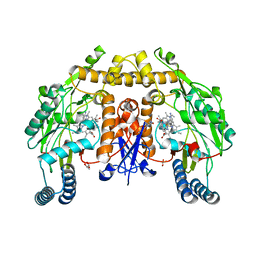 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)benzonitrile | | Descriptor: | 4-(2-{[(2-aminoquinolin-7-yl)methyl]amino}ethyl)benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
6CWX
 
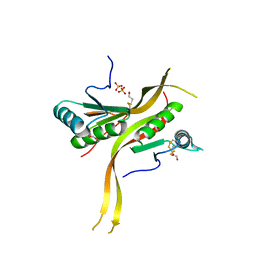 | | Crystal structure of human ribonuclease P/MRP proteins Rpp20/Rpp25 | | Descriptor: | FORMIC ACID, Ribonuclease P protein subunit p20, Ribonuclease P protein subunit p25, ... | | Authors: | Chan, C.W, Kiesel, B.R, Mondragon, A. | | Deposit date: | 2018-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rpp20/Rpp25 Reveals Quaternary Level Adaptation of the Alba Scaffold as Structural Basis for Single-stranded RNA Binding.
J. Mol. Biol., 430, 2018
|
|
6CI9
 
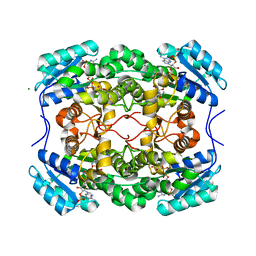 | |
4X6R
 
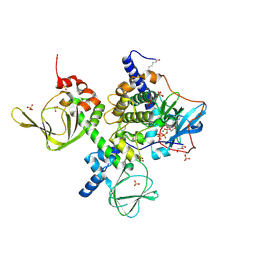 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Wu, J, Taylor, S.S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-07-22 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
4X7D
 
 | |
2O3Z
 
 | | X-ray crystal structure of LpxC complexed with 3-heptyloxybenzoate | | Descriptor: | 3-(heptyloxy)benzoic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Christianson, D.W. | | Deposit date: | 2006-12-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Amphipathic benzoic acid derivatives: synthesis and binding in the hydrophobic tunnel of the zinc deacetylase LpxC.
Bioorg.Med.Chem., 15, 2007
|
|
