1Q3K
 
 | |
2GKW
 
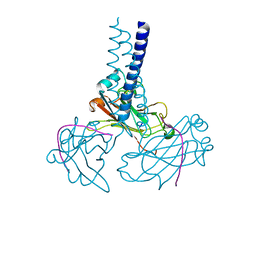 | | Key contacts promote recongnito of BAFF-R by TRAF3 | | Descriptor: | TNF receptor-associated factor 3, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Ely, K.R. | | Deposit date: | 2006-04-03 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Key molecular contacts promote recognition of the BAFF receptor by TNF receptor-associated factor 3: implications for intracellular signaling regulation.
J.Immunol., 173, 2004
|
|
403D
 
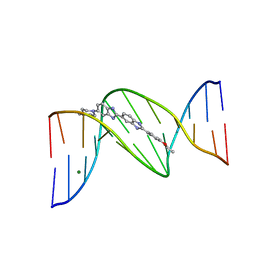 | | 5'-D(*CP*GP*CP*(HYD)AP*AP*AP*TP*TP*TP*GP*CP*G)-3', 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(IGU)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Bauer, C, Roberts, C, Switzer, C, Wang, A.H.-J. | | Deposit date: | 1998-06-10 | | Release date: | 1998-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis.
Biochemistry, 37, 1998
|
|
4Q4S
 
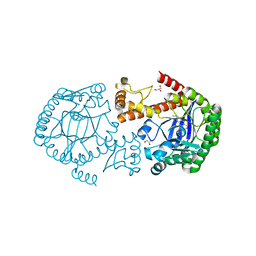 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-2-[(thiophen-2-ylmethyl)amino]-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(thiophen-2-ylmethyl)amino]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Beyond Affinity: Enthalpy-Entropy Factorization Unravels Complexity of a Flat Structure-Activity Relationship for Inhibition of a tRNA-Modifying Enzyme.
J.Med.Chem., 57, 2014
|
|
1J7S
 
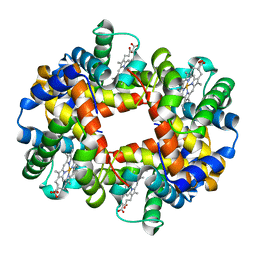 | | Crystal Structure of deoxy HbalphaYQ, a mutant of HbA | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
8ZTZ
 
 | | Structure of ATP-dependent diazotase CmaA6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Putative AMP-binding enzyme | | Authors: | Katsuyama, Y, Kawai, S, Ohnishi, Y. | | Deposit date: | 2024-06-07 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of ATP-Dependent Diazotase CmaA6.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1C4A
 
 | |
1C4C
 
 | |
3JTA
 
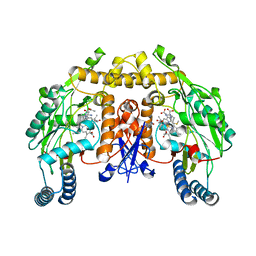 | | Structure of neuronal nitric oxide synthase heme domain in the ferrous state complexed with N~5~-[4-(methylsulfanyl)butanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
5ZH4
 
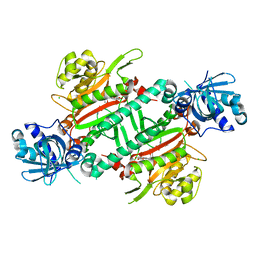 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-7 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2S,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
4RF7
 
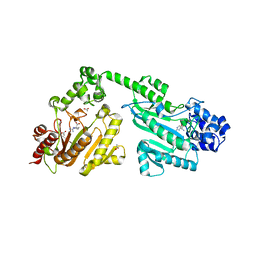 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with substrate L-arginine | | Descriptor: | ACETATE ION, ARGININE, Arginine kinase | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2AU4
 
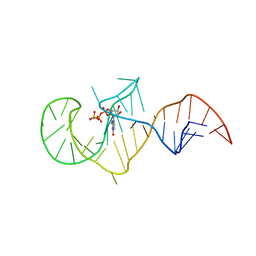 | | Class I GTP aptamer | | Descriptor: | Class I RNA aptamer to GTP, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Carothers, J.M, Davis, J.H, Chou, J.J, Szostak, J.W. | | Deposit date: | 2005-08-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an informationally complex high-affinity RNA aptamer to GTP.
Rna, 12, 2006
|
|
2GW2
 
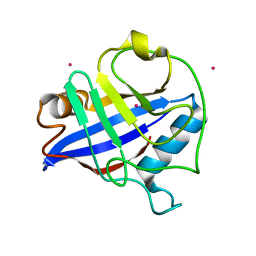 | | Crystal structure of the peptidyl-prolyl isomerase domain of human cyclophilin G | | Descriptor: | Peptidyl-prolyl cis-trans isomerase G, UNKNOWN ATOM OR ION | | Authors: | Bernstein, G, Tempel, W, Davis, T, Newman, E.M, Finerty Jr, P.J, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
4RQT
 
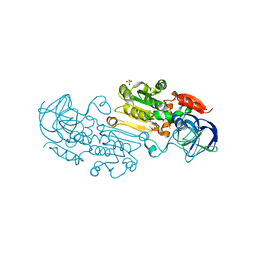 | | Alcohol Dehydrogenase Crystal Structure | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase class-P, SULFATE ION, ... | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
4OGW
 
 | | Structure of a human CD38 mutant complexed with NMN | | Descriptor: | ADP-ribosyl cyclase 1, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE | | Authors: | Shewchuk, L.M, Preugschat, F, Carter, L.H, Boros, E.E, Moyer, M.B, Stewart, E.L, Porter, D.J.T. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A pre-steady state and steady state kinetic analysis of the N-ribosyl hydrolase activity of hCD157.
Arch.Biochem.Biophys., 564C, 2014
|
|
4Q4R
 
 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 2-{[2-(Morpholin-4-yl)ethyl]amino}-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 2-{[2-(4-Morpholinyl)ethyl]amino}-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Beyond Affinity: Enthalpy-Entropy Factorization Unravels Complexity of a Flat Structure-Activity Relationship for Inhibition of a tRNA-Modifying Enzyme.
J.Med.Chem., 57, 2014
|
|
3JT8
 
 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-{3-[(1-methylethyl)sulfanyl]propanimidoyl}-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JT4
 
 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-[(3-(ethylsulfanyl)propanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
4QSI
 
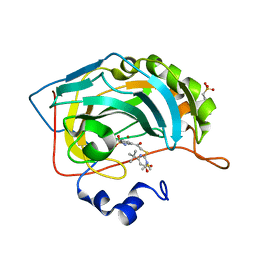 | | Crystal structure of human carbonic anhydrase isozyme II with 5-{[(4-tert-buthyl-6-oxo-1,6-dihydropyrimidin-2-yl)thio]acetyl}-2-chlorobenzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-{[(4-tert-butyl-6-oxo-1,6-dihydropyrimidin-2-yl)sulfanyl]acetyl}-2-chlorobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-07-04 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Intrinsic Thermodynamics and Structure Correlation of Benzenesulfonamides with a Pyrimidine Moiety Binding to Carbonic Anhydrases I, II, VII, XII, and XIII
Plos One, 9, 2014
|
|
1BKX
 
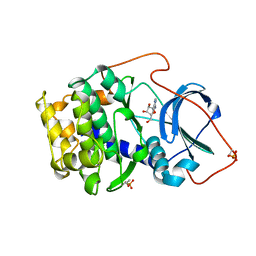 | | A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY | | Descriptor: | ADENOSINE MONOPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Cox, S, Xuong, N, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility.
Structure, 5, 1997
|
|
3ZO1
 
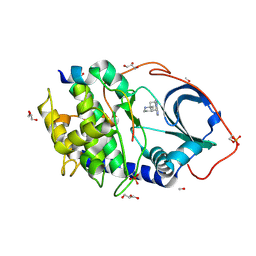 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | 6-(1,9-DIAZASPIRO[5.5]UNDECAN-9-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
4R64
 
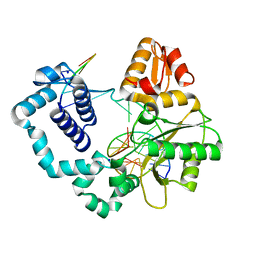 | | Binary complex crystal structure of E295K mutant of DNA polymerase Beta | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
4R66
 
 | | Ternary complex crystal structure of E295K mutant of DNA polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
4QT6
 
 | | Crystal structure of the SPRY domain of human HERC1 | | Descriptor: | FORMAMIDE, Probable E3 ubiquitin-protein ligase HERC1, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Guan, X, Wernimont, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2015-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the SPRY domain of human HERC1
To be Published
|
|
4C34
 
 | | PKA-S6K1 Chimera with Staurosporine bound | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, GLYCEROL, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
