2GFR
 
 | |
7WX6
 
 | | A Legionella acetyltransferase VipF | | Descriptor: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6P3K
 
 | | Crystal structure of LigU(C100S) | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
4TNL
 
 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6X9B
 
 | |
7DTA
 
 | |
6X99
 
 | |
6X9C
 
 | |
1A5T
 
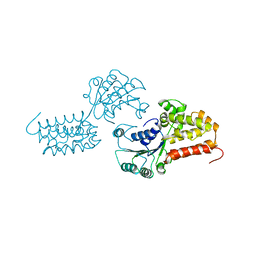 | | CRYSTAL STRUCTURE OF THE DELTA PRIME SUBUNIT OF THE CLAMP-LOADER COMPLEX OF ESCHERICHIA COLI DNA POLYMERASE III | | Descriptor: | DELTA PRIME, ZINC ION | | Authors: | Guenther, B, Onrust, R, Sali, A, O'Donnell, M, Kuriyan, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the delta' subunit of the clamp-loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 91, 1997
|
|
6X9A
 
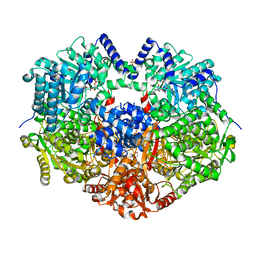 | |
6X9D
 
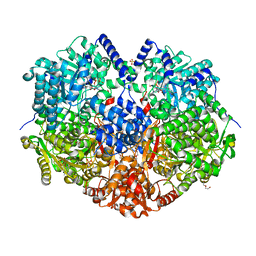 | |
1EJ9
 
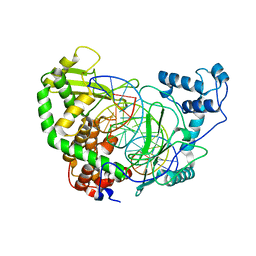 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
1FAQ
 
 | |
3L11
 
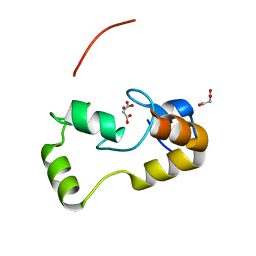 | | Crystal Structure of the Ring Domain of RNF168 | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MALONATE ION, ZINC ION | | Authors: | Neculai, D, Yermekbayeva, L, Crombet, L, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
8E5S
 
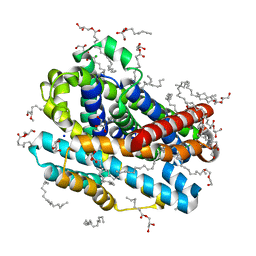 | |
8E6L
 
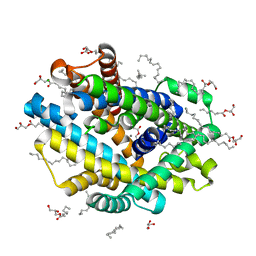 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter D296A mutant in an inward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Divalent metal cation transporter MntH, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E6H
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter A47W mutant in an occluded, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E6N
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter G223W mutant in an outward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Divalent metal cation transporter MntH, ... | | Authors: | Bozzi, A.T, Ray, S, Zimanyi, C.M, Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E6M
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter WT in an inward-open, cadmium-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E5V
 
 | |
8E60
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter WT in an occluded, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E6I
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter M230A mutant in an inward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
1FAR
 
 | |
7CPP
 
 | |
3NW5
 
 | | Crystal structure of insulin-like growth factor 1 receptor (IGF-1R-WT) complex with a carbon-linked proline isostere inhibitor (11B) | | Descriptor: | Insulin-like growth factor 1 receptor, N-(5-cyclopropyl-1H-pyrazol-3-yl)-2-{(2R)-1-[(6-fluoropyridin-3-yl)carbonyl]pyrrolidin-2-yl}pyrrolo[2,1-f][1,2,4]triazin-4-amine | | Authors: | Sack, J.S. | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Proline isosteres in a series of 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitors of IGF-1R kinase and IR kinase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
