5HF7
 
 | | TDG enzyme-substrate complex | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5HRY
 
 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5H5Y
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
3L6V
 
 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | Descriptor: | DNA gyrase subunit A | | Authors: | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | Deposit date: | 2009-12-26 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
5HT2
 
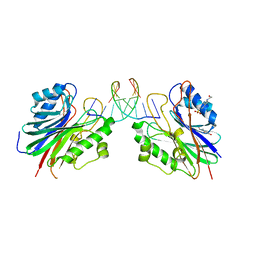 | | Mouse Tdp2 reaction product (5'-phosphorylated DNA)-Mg2+ complex with 1-N6-etheno-adenine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DNA, ... | | Authors: | Schellenberg, M.J, Vilas, C.K, Williams, R.S. | | Deposit date: | 2016-01-26 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2.
Nucleic Acids Res., 44, 2016
|
|
6OSR
 
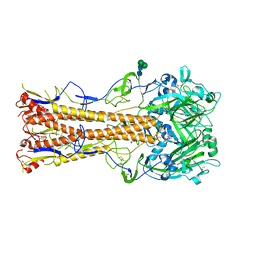 | |
3LLK
 
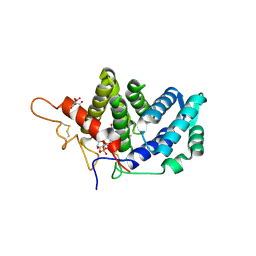 | | Sulfhydryl Oxidase Fragment of Human QSOX1 | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Sulfhydryl oxidase 1 | | Authors: | Alon, A, Fass, D. | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | QSOX contains a pseudo-dimer of functional and degenerate sulfhydryl oxidase domains.
Febs Lett., 584, 2010
|
|
5HU2
 
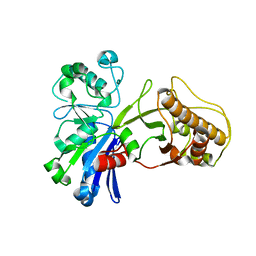 | |
3L7H
 
 | |
5H6L
 
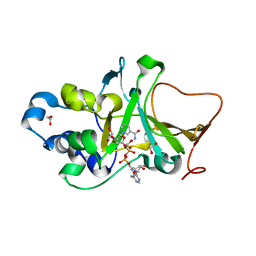 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6X
 
 | | The crystal structure of RpoS fragment including a partial region 1.2 and region 2 from intracellular pathogen Legionella pneumophila | | Descriptor: | GLYCEROL, RNA polymerase sigma factor RpoS, SODIUM ION | | Authors: | Zhang, N, Chen, X, Gong, X, Ge, H. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of RpoS Fragment including a Partial Region 1.2 and Region 2 from the Intracellular Pathogen Legionella pneumophila
Crystals, 8, 2018
|
|
5HUC
 
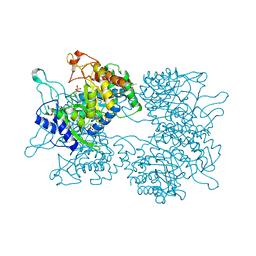 | | DAHP synthase from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, MANGANESE (II) ION, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
3L7P
 
 | |
5H71
 
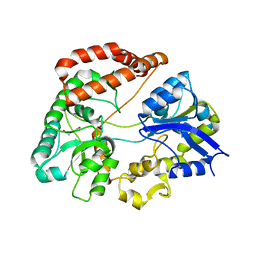 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate trisaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgQ2, CALCIUM ION, ... | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
3L7T
 
 | |
5HUN
 
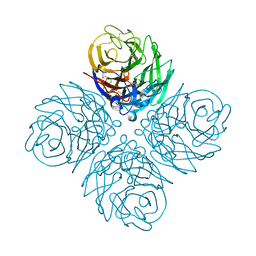 | | The crystal structure of neuraminidase from A/gyrfalcon/Washington/41088-6/2014 influenza virus | | Descriptor: | CALCIUM ION, Neuraminidase | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
5H7K
 
 | | Crystal structure of Elongation factor 2 GDP-form | | Descriptor: | Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
3LNH
 
 | |
5HUV
 
 | |
3LNT
 
 | |
5H7T
 
 | |
5H7W
 
 | |
3LOA
 
 | |
5HV8
 
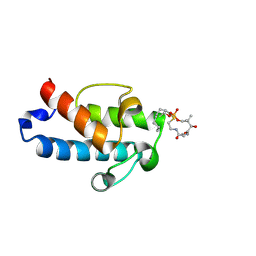 | | Solution structure of an octanoyl- loaded acyl carrier protein domain from module MLSA2 of the mycolactone polyketide synthase. | | Descriptor: | S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] octanethioate, Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
5HVX
 
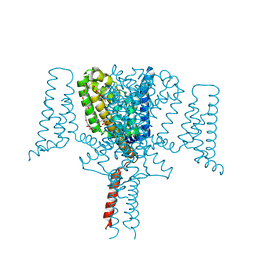 | | Full length Wild-Type Open-form Sodium Channel NavMs | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, ... | | Authors: | Sula, A, Booker, J.E, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2016-01-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The complete structure of an activated open sodium channel.
Nat Commun, 8, 2017
|
|
