1JBD
 
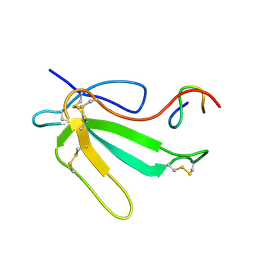 | | NMR Structure of the Complex Between alpha-bungarotoxin and a Mimotope of the Nicotinic Acetylcholine Receptor | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Klein, S, Niccolai, N. | | Deposit date: | 2001-06-04 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
7JL3
 
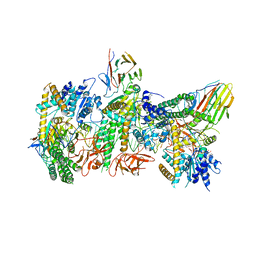 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JMD
 
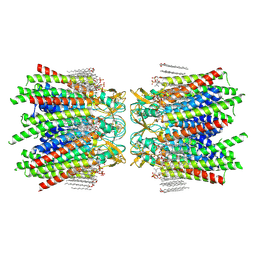 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JQ2
 
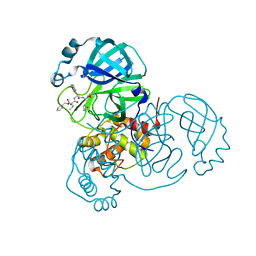 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI5 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
6C5T
 
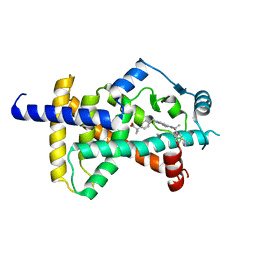 | | PPARg LBD bound to SR11023 | | Descriptor: | 2-{4-[(5-{[(1R)-1-(3-cyclopropylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]phenyl}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P.R. | | Deposit date: | 2018-01-16 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | PPAR gamma in Complex with an Antagonist and Inverse Agonist: a Tumble and Trap Mechanism of the Activation Helix.
iScience, 5, 2018
|
|
1H6D
 
 | |
6CCN
 
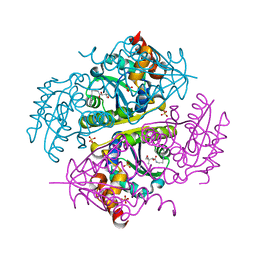 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-2,4-dihydroxy-N-(2-(4-hydroxy-1H-benzo[d]imidazol-2-yl)ethyl)-3,3-dimethylbutanamide | | Descriptor: | (2R)-2,4-dihydroxy-N-[2-(7-hydroxy-1H-benzimidazol-2-yl)ethyl]-3,3-dimethylbutanamide, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CUT
 
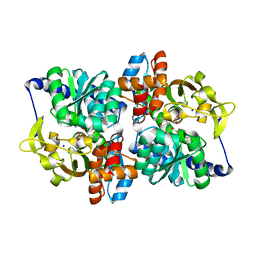 | | Engineered Holo TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3S)-isopropylserine bound as the external aldimine | | Descriptor: | (2S,3S)-3-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-methylpentanoic acid (non-preferred name), SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Boville, C.E, Scheele, R.A, Buller, A.R, Arnold, F.H. | | Deposit date: | 2018-03-26 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6CV7
 
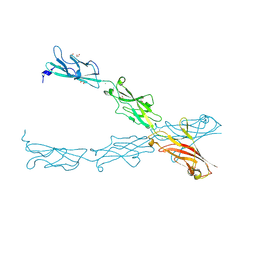 | |
1JH7
 
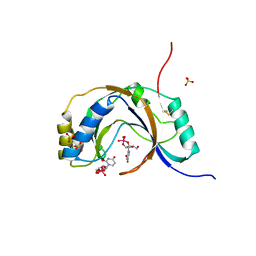 | | Semi-reduced Inhibitor-bound Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, URIDINE-2',3'-VANADATE, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
1JF7
 
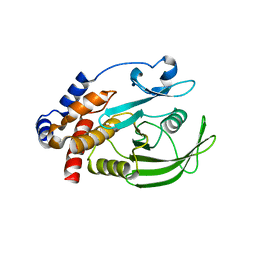 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
1JFK
 
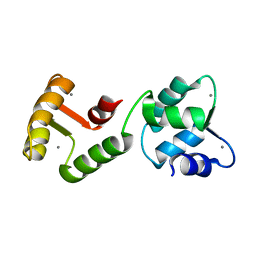 | | MINIMUM ENERGY REPRESENTATIVE STRUCTURE OF A CALCIUM BOUND EF-HAND PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
1GY8
 
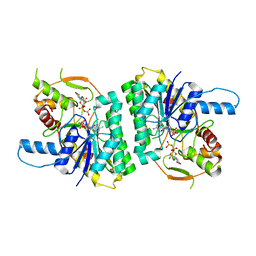 | | Trypanosoma brucei UDP-galactose 4' epimerase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Shaw, M.P, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-04-21 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Crystal Structure of Trypanosoma Brucei Udp-Galactose 4'-Epimerase: A Potential Target for Structure-Based Development of Novel Trypanocides
Mol.Biochem.Parasitol., 126, 2003
|
|
1JI6
 
 | |
1JAL
 
 | |
1H96
 
 | | recombinant mouse L-chain ferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, SULFATE ION | | Authors: | Granier, T, Gallois, B, D'Estaintot, B.L, Dautant, A, Chevalier, J.M, Mellado, J.M, Beaumont, C, Santambrogio, P, Arosio, P, Precigoux, G. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mouse L-Chain Ferritin at 1.6 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GPQ
 
 | | Structure of ivy complexed with its target, HEWL | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Monchois, V, Claverie, J.-M. | | Deposit date: | 2001-11-08 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6CI9
 
 | |
7GGY
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-966f8da6-2 (Mpro-x12717) | | Descriptor: | (4R)-1-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1JDB
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Wesenberg, G, Raushel, F.M, Rayment, I. | | Deposit date: | 1997-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1JJJ
 
 | | SOLUTION STRUCTURE OF RECOMBINANT HUMAN EPIDERMAL-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | EPIDERMAL-TYPE FATTY ACID BINDING PROTEIN (E-FABP) | | Authors: | Gutierrez-Gonzalez, L.H, Ludwig, C, Hohoff, C, Rademacher, M, Hanhoff, T, Rueterjans, H, Spener, F, Luecke, C. | | Deposit date: | 2001-07-06 | | Release date: | 2002-06-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human epidermal-type fatty
acid-binding protein (E-FABP)
BIOCHEM.J., 364, 2002
|
|
6CKM
 
 | |
6D1O
 
 | | FT_5 dioxygenase apoenzyme | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
7GCC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-95b75b4d-4 (Mpro-x10566) | | Descriptor: | 2-(3-cyclopropylphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1HA7
 
 | | STRUCTURE OF A LIGHT-HARVESTING PHYCOBILIPROTEIN, C-PHYCOCYANIN FROM SPIRULINA PLATENSIS AT 2.2A RESOLUTION | | Descriptor: | C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, PHYCOCYANOBILIN | | Authors: | Padyana, A.K, Rajashankar, K.R, Ramakumar, S. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Light-Harvesting Protein C-Phycocyanin from Spirulina Platensis
Biochem.Biophys.Res.Commun., 282, 2001
|
|
