5J33
 
 | | Isopropylmalate dehydrogenase in complex with NAD+ | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
1TQS
 
 | | Golgi alpha-Mannosidase II In Complex With Salacinol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIDEOXY-1,4-[[2S,3S)-2,4-DIHYDROXY-3-(SULFOXY)BUTYL]EPISULFONIUMYLIDENE]-D-ARABINITOL INNER SALT, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Ghavami, A, Johnston, B.D, Pinto, B.M, Rose, D.R. | | Deposit date: | 2004-06-18 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis of the interactions of Drosophila melanogaster Golgi alpha-mannosidase II with the naturally occurring glycomimetic salacinol and its analogues
Tetrahedron Asymmetry, 16, 2005
|
|
1H1Y
 
 | |
3NQB
 
 | |
5J5Z
 
 | | Crystal structure of the D444V disease-causing mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Mizsei, R, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2016-04-04 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Free Radic. Biol. Med., 124, 2018
|
|
1GQI
 
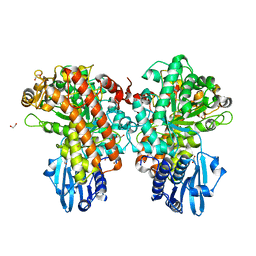 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
5IZ4
 
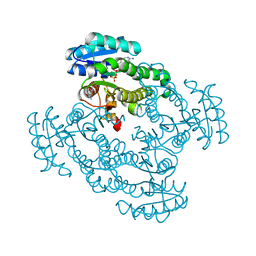 | |
1GUZ
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
5R8D
 
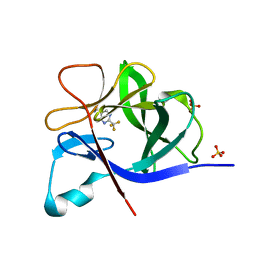 | |
5R8V
 
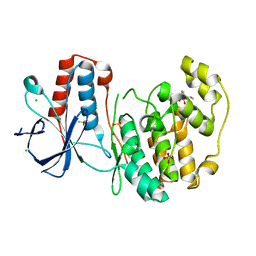 | |
5ITX
 
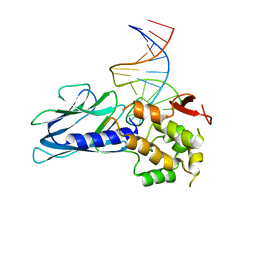 | | Crystal Structure of Human NEIL1(P2G R242K) bound to duplex DNA containing Thymine Glycol | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7EA4
 
 | |
1H09
 
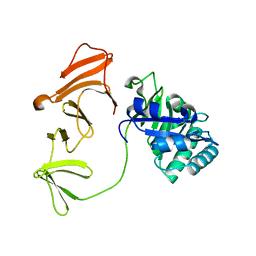 | | Multimodular Pneumococcal Cell Wall Endolysin from phage Cp-1 | | Descriptor: | LYSOZYME | | Authors: | Hermoso, J.A, Monterroso, B, Albert, A, Garcia, P, Menendez, M, Martinez-Ripoll, M, Garcia, J.L. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Selective Recognition of Pneumococcal Cell Wall by Modular Endolysin from Phage Cp-1
Structure, 11, 2003
|
|
7EBY
 
 | |
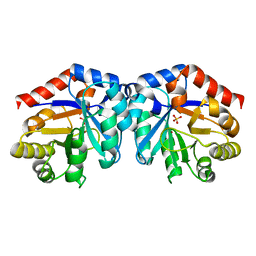
1H1Z
 
 | |
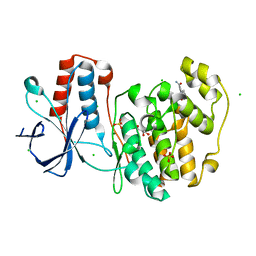
5R9A
 
 | |
4YUU
 
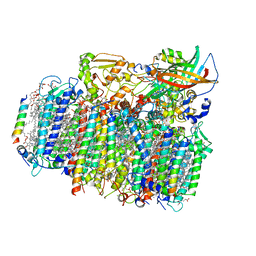 | | Crystal structure of oxygen-evolving photosystem II from a red alga | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ago, H, Shen, J.-R. | | Deposit date: | 2015-03-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7700038 Å) | | Cite: | Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
J.Biol.Chem., 291, 2016
|
|
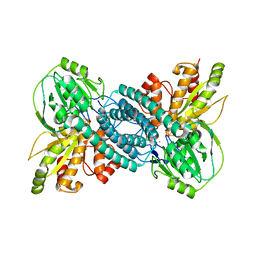
7E0L
 
 | |
5R9Q
 
 | |
7E4D
 
 | | Crystal structure of PlDBR | | Descriptor: | Double Bond Reductase | | Authors: | Sugimoto, K, Senda, M, Senda, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
5J55
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5RA5
 
 | |
4MUQ
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.364 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N9R
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentacholrocarbonyliridate(III) (1 day) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-10-21 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
1GRF
 
 | |
