1FBB
 
 | |
1PIN
 
 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
1FCU
 
 | | CRYSTAL STRUCTURE (TRIGONAL) OF BEE VENOM HYALURONIDASE | | Descriptor: | HYALURONOGLUCOSAMINIDASE | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Rizkallah, P.J, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
8J2V
 
 | |
8J74
 
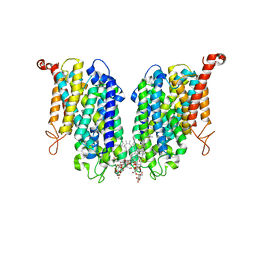 | | Human high-affinity choline transporter CHT1 in the HC-3-bound outward-facing open conformation, dimeric state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), CHOLESTEROL HEMISUCCINATE, HEXADECANE, ... | | Authors: | Gao, Y, Qiu, Y, Zhao, Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transport mechanism of presynaptic high-affinity choline uptake by CHT1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3A6Q
 
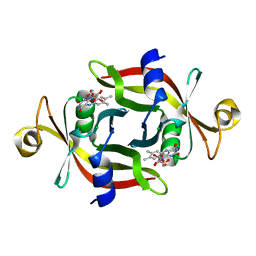 | | E13T mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
1FKP
 
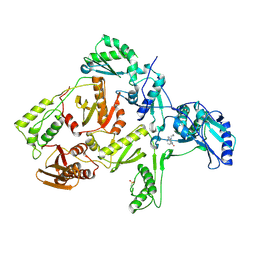 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
8J2U
 
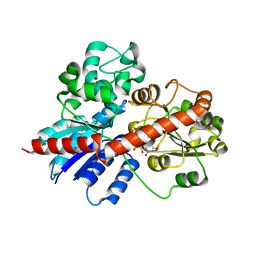 | |
8J5N
 
 | |
1FFA
 
 | |
8J31
 
 | |
1FA2
 
 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Lee, B.I, Cheong, C.G, Suh, S.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
1FAK
 
 | | HUMAN TISSUE FACTOR COMPLEXED WITH COAGULATION FACTOR VIIA INHIBITED WITH A BPTI-MUTANT | | Descriptor: | CALCIUM ION, PROTEIN (5L15), PROTEIN (BLOOD COAGULATION FACTOR VIIA), ... | | Authors: | Zhang, E, St Charles, R, Tulinsky, A. | | Deposit date: | 1998-12-28 | | Release date: | 1999-12-03 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of extracellular tissue factor complexed with factor VIIa inhibited with a BPTI mutant.
J.Mol.Biol., 285, 1999
|
|
8IO4
 
 | | Herg1a-herg1b open state | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
8J2Z
 
 | |
1PRT
 
 | | THE CRYSTAL STRUCTURE OF PERTUSSIS TOXIN | | Descriptor: | PERTUSSIS TOXIN (SUBUNIT S1), PERTUSSIS TOXIN (SUBUNIT S2), PERTUSSIS TOXIN (SUBUNIT S3), ... | | Authors: | Stein, P.E, Read, R.J. | | Deposit date: | 1993-11-22 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of pertussis toxin.
Structure, 2, 1994
|
|
1FFT
 
 | | The structure of ubiquinol oxidase from Escherichia coli | | Descriptor: | COPPER (II) ION, HEME O, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abramson, J, Riistama, S, Larsson, G, Jasaitis, A, Svensson-Ek, M, Puustinen, A, Iwata, S, Wikstrom, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site.
Nat.Struct.Biol., 7, 2000
|
|
1PVC
 
 | |
8J57
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in complex with bedaquiline(BDQ) | | Descriptor: | ATP synthase subunit a, ATP synthase subunit c, Bedaquiline | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
1FGU
 
 | |
8J7H
 
 | | ion channel | | Descriptor: | ILE-ALA-ALA-ILE-HIS-ASN-ALA-ARG-ARG-LYS-LYS-ARG-GLU-ALA-ALA-ALA-ALA-HIS-LYS-ALA, ion channel | | Authors: | Chen, H.W, Jiang, D.H. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | ion channel
To Be Published
|
|
8J6O
 
 | | transport T2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein (Fragment),SID1 transmembrane family member 2, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1Q99
 
 | | Crystal structure of the Saccharomyces cerevisiae SR protein kinsae, Sky1p, complexed with the non-hydrolyzable ATP analogue, AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, NICKEL (II) ION, ... | | Authors: | Nolen, B, Ngo, J, Chakrabarti, S, Vu, D, Adams, J.A, Ghosh, G. | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Nucleotide-Induced Conformational Changes in the Saccharomyces cerevisiae SR Protein Kinase, Sky1p, Revealed by X-ray Crystallography
Biochemistry, 42, 2003
|
|
1FID
 
 | |
8J9C
 
 | | Crystal structure of M61 peptidase (apo-form) from Xanthomonas campestris | | Descriptor: | GLYCEROL, Putative glycyl aminopeptidase, SODIUM ION, ... | | Authors: | Yadav, P, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
