3T6C
 
 | | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-gluconic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg
to be published
|
|
3TJ1
 
 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
3TOT
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 | | Descriptor: | ACETATE ION, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3TTE
 
 | | Crystal structure of enolase brado_4202 (target EFI-501651) from Bradyrhizobium complexed with magnesium and mandelic acid | | Descriptor: | (S)-MANDELIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mandelate Racemase from Bradyrhizobium Sp. Ors278
To be Published
|
|
4QJ8
 
 | |
3RHG
 
 | | Crystal structure of amidohydrolase pmi1525 (target efi-500319) from proteus mirabilis hi4320 | | Descriptor: | BENZOIC ACID, CACODYLATE ION, Putative phophotriesterase, ... | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
3U4R
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3UAP
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501774) from methylococcus capsulatus str. bath | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Methylococcus Capsulatus
To be Published
|
|
3UBK
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
3TOU
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 with GSH bound | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3QBT
 
 | | Crystal structure of OCRL1 540-678 in complex with Rab8a:GppNHp | | Descriptor: | Inositol polyphosphate 5-phosphatase OCRL-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hou, X, Hagemann, N, Schoebel, S, Blankenfeldt, W, Goody, R.S, Erdmann, K.S, Itzen, A. | | Deposit date: | 2011-01-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1.
Embo J., 30, 2011
|
|
3U4O
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TOY
 
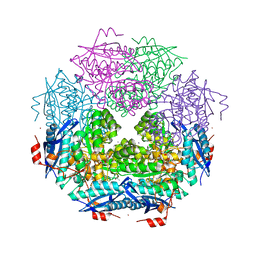 | | CRYSTAL STRUCTURE OF ENOLASE BRADO_4202 (TARGET EFI-501651) FROM Bradyrhizobium sp. ORS278 WITH CALCIUM AND ACETATE BOUND | | Descriptor: | ACETATE ION, CALCIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE FROM Bradyrhizobium sp. ORS278
To be Published
|
|
3TNF
 
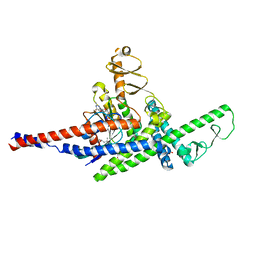 | | LidA from Legionella in complex with active Rab8a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LidA, MAGNESIUM ION, ... | | Authors: | Schoebel, S, Cichy, A.L, Goody, R.S, Itzen, A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein LidA from Legionella is a Rab GTPase supereffector.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TWB
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and gluconic acid | | Descriptor: | CHLORIDE ION, D-gluconic acid, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3TWA
 
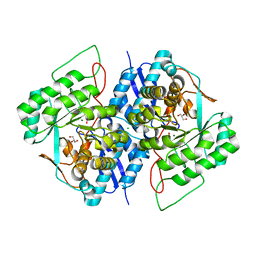 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3TO8
 
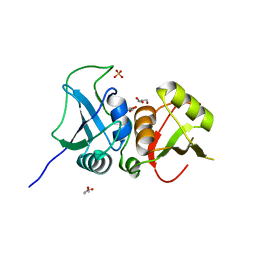 | | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Heterogeneous nuclear ribonucleoprotein L, ... | | Authors: | Zhang, W.J, Zeng, F.X, Liu, Y.W, Zhao, Y, Niu, L.W, Teng, M.K, Li, X. | | Deposit date: | 2011-09-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L)
To be Published
|
|
3TW9
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative dehydratase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3UAR
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501774) from methylococcus capsulatus str. bath with gsh bound | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Methylococcus Capsulatus
To be Published
|
|
3UBL
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans with gsh bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
5HIQ
 
 | |
5HIZ
 
 | | The structure of PEDV NSP9 | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
6E1F
 
 | |
5HIY
 
 | | Crystal structure of PEDV NSP9 Mutant-C59A | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
8TBP
 
 | | HLA-DRB1*15:01 in complex with smith antigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ting, Y.T, Broury, A, Ooi, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.12621117 Å) | | Cite: | Smith-specific regulatory T cells halt the progression of lupus nephritis.
Nat Commun, 15, 2024
|
|
