8IYL
 
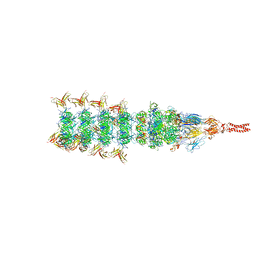 | | Tail tip conformation 2 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
3LP3
 
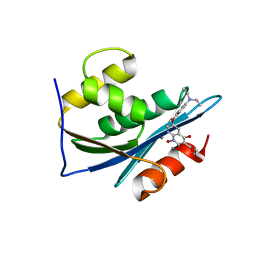 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
4EX6
 
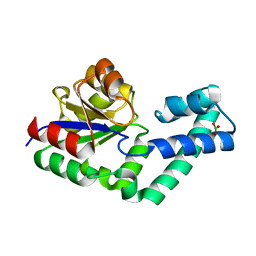 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7PE9
 
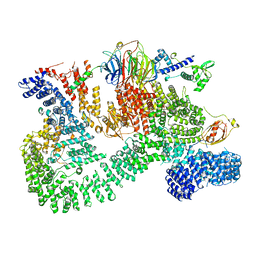 | |
7PEB
 
 | |
7PE8
 
 | |
7PEC
 
 | |
7PEA
 
 | |
7PE7
 
 | |
7PJ1
 
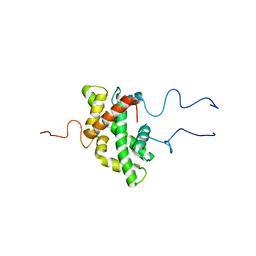 | |
7B5N
 
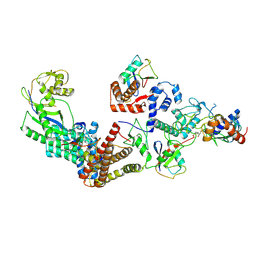 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-UBE2L3~Ub~ARIH1. | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, E3 ubiquitin-protein ligase ARIH1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
5MC6
 
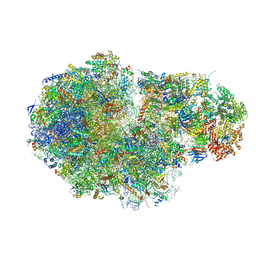 | | Cryo-EM structure of a native ribosome-Ski2-Ski3-Ski8 complex from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Schmidt, C, Kowalinski, E, Shanmuganathan, V, Defenouillere, Q, Braunger, K, Heuer, A, Pech, M, Namane, A, Berninghausen, O, Fromont-Racine, M, Jacquier, A, Conti, E, Becker, T, Beckmann, R. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex.
Science, 354, 2016
|
|
6GGD
 
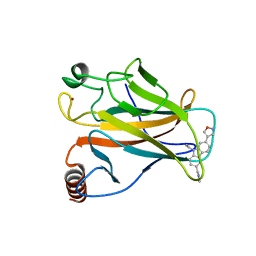 | |
6GGB
 
 | |
2VG6
 
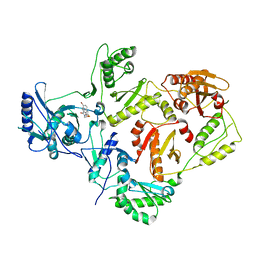 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
3NBH
 
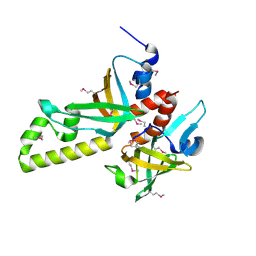 | | Crystal structure of human RMI1C-RMI2 complex | | Descriptor: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
6GGA
 
 | |
6TTU
 
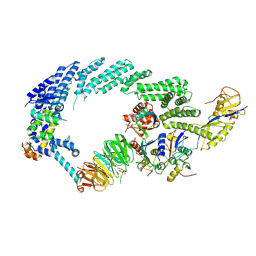 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
6GGE
 
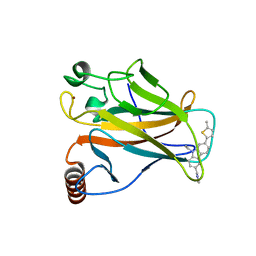 | | p53 cancer mutant Y220C in complex with small-molecule stabilizer PK9327 | | Descriptor: | Cellular tumor antigen p53, ZINC ION, [9-ethyl-7-(5-methylthiophen-2-yl)carbazol-3-yl]methyl-methyl-azanium | | Authors: | Joerger, A.C, Bauer, M.R. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25000513 Å) | | Cite: | A structure-guided molecular chaperone approach for restoring the transcriptional activity of the p53 cancer mutant Y220C.
Future Med Chem, 11, 2019
|
|
6GGC
 
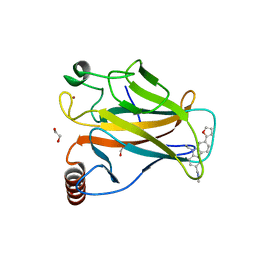 | |
2X6O
 
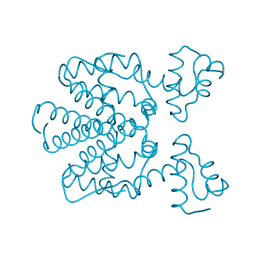 | | Tet Repressor class D in complex with 7-chlor-2-cyano-iso- tetracycline | | Descriptor: | (4S,4AS,6S,8AS)-6-[(1S)-7-CHLORO-4-HYDROXY-1-METHYL-3-OXO-1,3-DIHYDRO-2-BENZOFURAN-1-YL]-4-(DIMETHYLAMINO)-3,8A-DIHYDROXY-1,8-DIOXO-1,4,4A,5,6,7,8,8A-OCTAHYDRONAPHTHALENE-2-CARBONITRILE, CHLORIDE ION, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Volkers, G, Hinrichs, W. | | Deposit date: | 2010-02-18 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of Drug Degradation Products by Target Proteins: Isotetracycline Binding to Tet Repressor.
J.Med.Chem., 54, 2011
|
|
7KUA
 
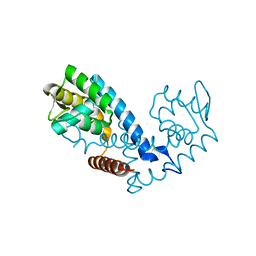 | | Crystal structure of the MarR family transcriptional regulator from Pseudomonas putida bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Transcriptional regulator, MarR family | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
2VG5
 
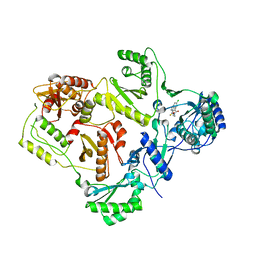 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
