4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
1BBH
 
 | |
3GYX
 
 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
2MKP
 
 | | N domain of cardiac troponin C bound to the switch fragment of fast skeletal troponin I at pH 6 | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Robertson, I.M, Pineda-Sanabria, S.E, Holmes, P.C, Sykes, B.D. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformation of the critical pH sensitive region of troponin depends upon a single residue in troponin I.
Arch.Biochem.Biophys., 552-553, 2014
|
|
4M6X
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M73
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2R,3R)-2-hydroxy-3-methoxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M74
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2S,3R)-2,3-dihydroxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2JJR
 
 | | V232K, N236D-trichosanthin | | Descriptor: | DI(HYDROXYETHYL)ETHER, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN, SULFATE ION, ... | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
4Q96
 
 | | CID of human RPRD1B in complex with an unmodified CTD peptide | | Descriptor: | RPB1-CTD, Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7CJX
 
 | | UDP-glucuronosyltransferase 2B15 C-terminal domain-L446S | | Descriptor: | L(+)-TARTARIC ACID, UDP-glucuronosyltransferase 2B15 | | Authors: | Wang, C.Y, Zhang, L. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.986414 Å) | | Cite: | Structure of UDP-glucuronosyltransferase 2B15 C-terminal domain L446S at 1.99 Angstroms resolution
To Be Published
|
|
4MJN
 
 | |
4M6Y
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M72
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2R,3R)-2-hydroxy-3-methoxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1I8O
 
 | |
1OB1
 
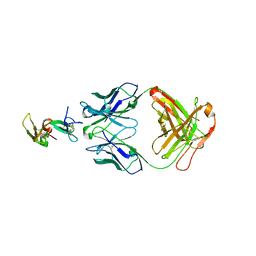 | | Crystal structure of a Fab complex whith Plasmodium falciparum MSP1-19 | | Descriptor: | ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pizarro, J.C, Chitarra, V, Verger, D, Holm, I, Petres, S, Dartville, S, Nato, F, Longacre, S, Bentley, G.A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Fab Complex Formed with Pfmsp1-19, the C-Terminal Fragment of Merozoite Surface Protein 1 from Plasmodium Falciparum: A Malaria Vaccine Candidate
J.Mol.Biol., 328, 2003
|
|
1UO9
 
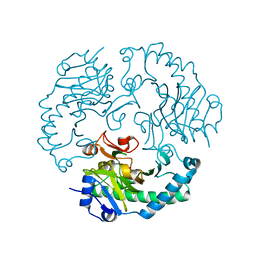 | | Deacetoxycephalosporin C synthase complexed with succinate | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, SUCCINIC ACID | | Authors: | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
4P5H
 
 | |
1OAE
 
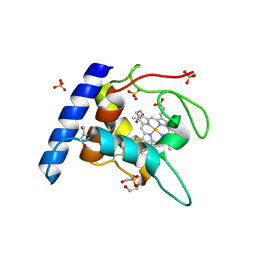 | | Crystal structure of the reduced form of cytochrome c" from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C", GLYCEROL, HEME C, ... | | Authors: | Enguita, F.J, Grenha, R, Santos, H, Carrondo, M.A. | | Deposit date: | 2003-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
2JUJ
 
 | |
1CYF
 
 | |
1CSJ
 
 | | CRYSTAL STRUCTURE OF THE RNA-DEPENDENT RNA POLYMERASE OF HEPATITIS C VIRUS | | Descriptor: | HEPATITIS C VIRUS RNA POLYMERASE (NS5B) | | Authors: | Bressanelli, S, Tomei, L, Roussel, A, Incitti, I, Vitale, R.L, Mathieu, M, De Francesco, R, Rey, F.A. | | Deposit date: | 1999-08-18 | | Release date: | 1999-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3UBR
 
 | | Laue structure of Shewanella oneidensis cytochrome-c Nitrite Reductase | | Descriptor: | CALCIUM ION, Cytochrome c-552, HEME C | | Authors: | Youngblut, M, Judd, E.T, Srajer, V, Sayed, B, Goeltzner, T, Elliott, S, Schmidt, M, Pacheco, A. | | Deposit date: | 2011-10-24 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system.
J.Biol.Inorg.Chem., 17, 2012
|
|
7EI4
 
 | | Crystal structure of MasL in complex with a novel covalent inhibitor, collimonin C | | Descriptor: | (6S,7R,9E)-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
4QDR
 
 | |
4F08
 
 | |
