1GBX
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1FZ2
 
 | |
5JEN
 
 | |
4YLW
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, D, Ouyang, P, Lu, W, Huang, J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound
To Be Published
|
|
4L4F
 
 | | Structure of cyanide and camphor bound P450cam mutant L358A/K178G/D182N | | Descriptor: | CAMPHOR, CYANIDE ION, Camphor 5-monooxygenase, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
5JGU
 
 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5JFL
 
 | |
4LAF
 
 | |
1S0T
 
 | | Solution structure of a DNA duplex containing an alpha-anomeric adenosine: insights into substrate recognition by endonuclease IV | | Descriptor: | 5'-D(*Cp*Gp*Tp*Cp*Gp*Tp*Gp*Gp*Ap*C)-3', 5'-D(*Gp*Tp*Cp*Cp*(A3A)p*Cp*Gp*Ap*Cp*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-04 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
4YNI
 
 | |
7F1N
 
 | | Beta-Glucosidase | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Anke, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14000368 Å) | | Cite: | Structural and Catalytic Characterization of TsBGL, a beta-Glucosidase From Thermofilum sp. ex4484_79.
Front Microbiol, 12, 2021
|
|
1G79
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI PYRIDOXINE 5'-PHOSPHATE OXIDASE COMPLEXED WITH PYRIDOXAL 5'-PHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Safo, M.K, Musayev, F.N, di Salvo, M.L, Schirch, V. | | Deposit date: | 2000-11-09 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Escherichia coli pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 2.0 A resolution.
J.Mol.Biol., 310, 2001
|
|
4YSY
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[(2,4-dichlorophenyl)methyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
3O38
 
 | |
3OE0
 
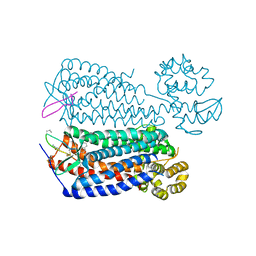 | | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 | | Descriptor: | C-X-C chemokine receptor type 4, Lysozyme Chimera, Polyphemusin analog, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
5JIF
 
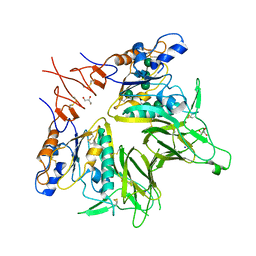 | | Crystal structure of mouse hepatitis virus strain DVIM Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4LCR
 
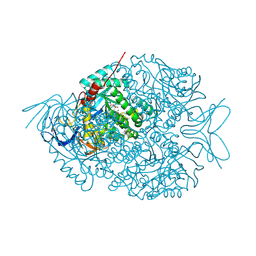 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
3OEC
 
 | |
1G92
 
 | | SOLUTION STRUCTURE OF PONERATOXIN | | Descriptor: | PONERATOXIN | | Authors: | Szolajska, E, Poznanski, J, Ferber, M.L, Michalik, J, Gout, E, Fender, P, Bailly, I, Dublet, B, Chroboczek, J. | | Deposit date: | 2000-11-22 | | Release date: | 2003-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Poneratoxin, a neurotoxin from ant venom. Structure and expression in insect cells and construction of a bio-insecticide.
Eur.J.Biochem., 271, 2004
|
|
1FZ7
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM III SOAKED IN 0.9 M ETHANOL | | Descriptor: | CALCIUM ION, ETHANOL, FE (III) ION, ... | | Authors: | Whittington, D.A, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase.
J.Am.Chem.Soc., 123, 2001
|
|
1GAZ
 
 | | Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues at turns in the conformational stability and folding of human lysozyme.
Biochemistry, 39, 2000
|
|
3ODU
 
 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
5JK6
 
 | |
4LA4
 
 | |
3OC3
 
 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
