4QPN
 
 | | Crystal Structure of Human Methyltransferase-Like Protein 21B | | Descriptor: | Protein-lysine methyltransferase METTL21B, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Tempel, W, Hong, B.S, Seitova, A, He, H, Li, Y, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Human Methyltransferase-Like Protein 21B
TO BE PUBLISHED
|
|
7XXC
 
 | | Orf1-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4XHE
 
 | | Crystal Structure of A-AChBP in complex with pinnatoxin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pinnatoxin A, ... | | Authors: | Bourne, Y, Sulzenbacher, G, Marchot, P. | | Deposit date: | 2015-01-05 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Marine Macrocyclic Imines, Pinnatoxins A and G: Structural Determinants and Functional Properties to Distinguish Neuronal alpha 7 from Muscle alpha 12 beta gamma delta nAChRs.
Structure, 23, 2015
|
|
4QV8
 
 | | yCP beta5-C52F mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QVV
 
 | | yCP beta5-A49V mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QWS
 
 | | yCP beta5-C63F mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QZ6
 
 | | yCP beta5-A49T-A50V double mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7XVV
 
 | |
7XVW
 
 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5BNZ
 
 | |
7XXM
 
 | | Orf1-glycine-4-aminobutylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4RG7
 
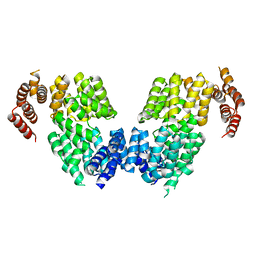 | | Crystal structure of APC3 | | Descriptor: | Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
7XX0
 
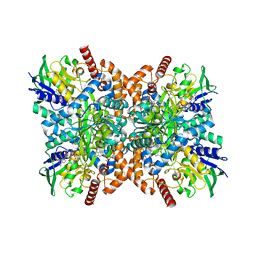 | | C281S glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XYL
 
 | | E426Q-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
6M4G
 
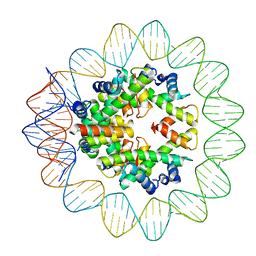 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
3U58
 
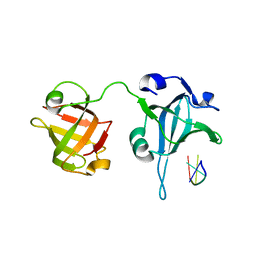 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 AB | | Descriptor: | DNA (5'-D(*GP*GP*GP*T)-3'), Tetrahymena Teb1 AB | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8QBY
 
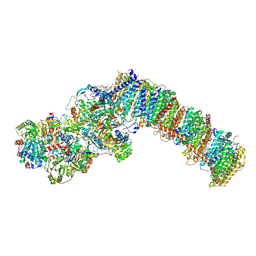 | | Respiratory complex I from Paracoccus denitrificans in MSP2N2 nanodiscs | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Ivanov, B.S, Bridges, H.R, Hirst, J. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Respiratory complex I from Paracoccus denitrificans
To Be Published
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4QV6
 
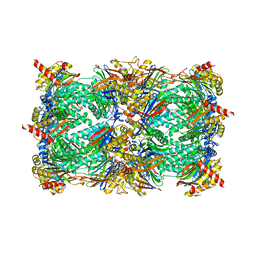 | | yCP beta5-A49V mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QW7
 
 | | yCP beta5-M45T mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7XXD
 
 | | Orf1-sarcosine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, SARCOSINE | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4QXJ
 
 | | yCP beta5-M45A mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7Y0X
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4QZW
 
 | | yCP beta5-C52F mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7XQA
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
