3ID4
 
 | | Crystal Structure of RseP PDZ2 domain fused GKASPV peptide | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID3
 
 | | Crystal Structure of RseP PDZ2 I304A domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6PTU
 
 | |
5WIB
 
 | | Structure of Acinetobacter baumannii carbapenemase OXA-239 K82D bound to imipenem | | Descriptor: | Imipenem, OXA-239 | | Authors: | Harper, T.M, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2017-07-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239.
Biochem. J., 475, 2018
|
|
6WMQ
 
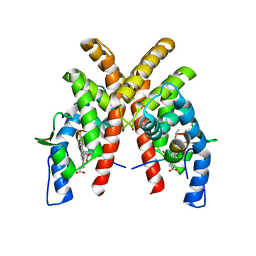 | |
6N6U
 
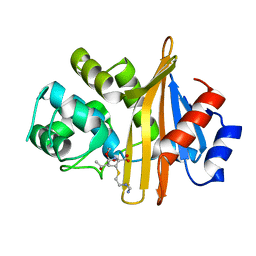 | |
5EPH
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 in complex with Imipenem | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Pozzi, C, Benvenuti, M, De Luca, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
8HIP
 
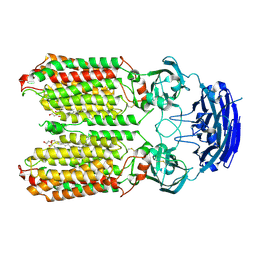 | | dsRNA transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Systemic RNA interference defective protein 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HKE
 
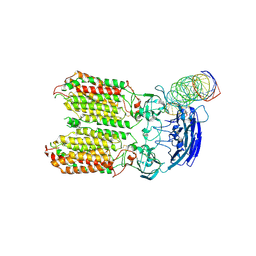 | | dsRNA transporter | | Descriptor: | RNA (37-MER), Systemic RNA interference defective protein 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3I67
 
 | | Ribonuclease A by LB nanotemplate method after high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3I6H
 
 | | Ribonuclease A by LB nanotemplate method before high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3I30
 
 | | Proteinase K by Classical hanging drop Method after high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To be Published
|
|
3I2Y
 
 | | Proteinase K by Classical hanging drop Method before high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.995 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To be Published
|
|
3I37
 
 | | Proteinase K by LB Nanotemplate Method before high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.995 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To Be Published
|
|
3I6F
 
 | | Ribonuclease A by Classical hanging drop method before high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3DVQ
 
 | |
3DW1
 
 | |
3DVS
 
 | |
3DO2
 
 | |
3I6J
 
 | | Ribonuclease A by Classical hanging drop method after high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
6BH0
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (R)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N51) | | Descriptor: | 2-{(R)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGZ
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methyl-1H-imidazol-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N47) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(1-methyl-1H-imidazol-2-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGV
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGY
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
