1YX9
 
 | |
1SLU
 
 | |
1YZE
 
 | | Crystal structure of the N-terminal domain of USP7/HAUSP. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M.N, Shire, K, Nguyen, T, Zhang, R.G, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1S8D
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-3A | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-02-02 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
2B2U
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
1V6Z
 
 | |
1ZCP
 
 | |
1V7X
 
 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1Z36
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z3D
 
 | | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2 1 | | Authors: | Gavira, J.A, DiGiammarino, E, Tempel, W, Toh, D, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans
To be Published
|
|
2B4C
 
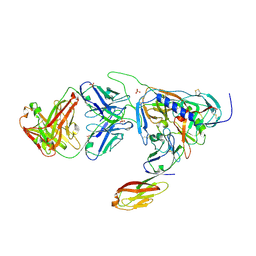 | | Crystal structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) complexed with CD4 and the X5 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, T-cell surface glycoprotein CD4, ... | | Authors: | Huang, C, Tang, M, Zhang, M.Y, Majeed, S, Montabana, E, Stanfield, R.L, Dimitrov, D.S, Korber, B, Sodroski, J, Wilson, I.A, Wyatt, R, Kwong, P.D. | | Deposit date: | 2005-09-23 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a V3-containing HIV-1 gp120 core.
Science, 310, 2005
|
|
1ZG8
 
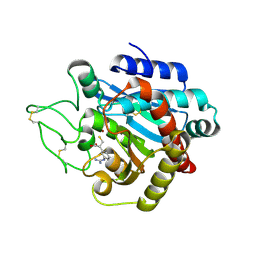 | | Crystal Structure of (R)-2-(3-{[amino(imino)methyl]amino}phenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | (2R)-2-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1VAL
 
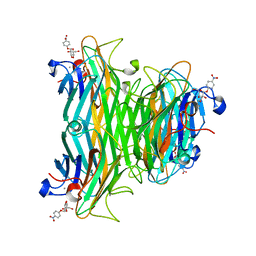 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
1ZJ5
 
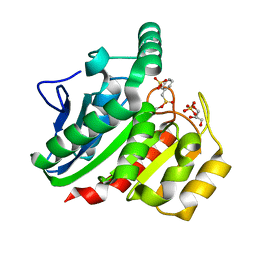 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1GQ8
 
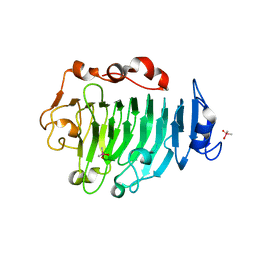 | | Pectin methylesterase from Carrot | | Descriptor: | CACODYLATE ION, PECTINESTERASE | | Authors: | Johansson, K, El-Ahmad, M, Friemann, R, Jornvall, H, Markovic, O, Eklund, H. | | Deposit date: | 2001-11-20 | | Release date: | 2002-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Plant Pectin Methylesterase
FEBS Lett., 514, 2002
|
|
2B7J
 
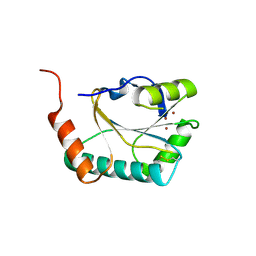 | |
1URO
 
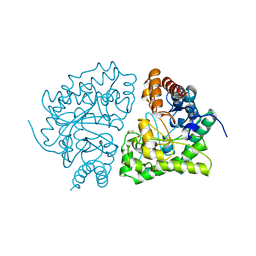 | | UROPORPHYRINOGEN DECARBOXYLASE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (UROPORPHYRINOGEN DECARBOXYLASE) | | Authors: | Whitby, F.G, Phillips, J.D, Kushner, J.P, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human uroporphyrinogen decarboxylase.
EMBO J., 17, 1998
|
|
1H64
 
 | |
1V73
 
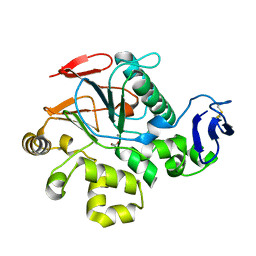 | | Crystal Structure of Cold-Active Protein-Tyrosine Phosphatase of a Psychrophile Shewanella SP. | | Descriptor: | ACETIC ACID, CALCIUM ION, psychrophilic phosphatase I | | Authors: | Tsuruta, H, Mikami, B, Aizono, Y. | | Deposit date: | 2003-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of Cold-Active Protein-Tyrosine Phosphatase from a Psychrophile, Shewanella sp
J.Biochem.(Tokyo), 137, 2005
|
|
2BBB
 
 | | Structure of HIV1 protease and hh1_173_3a complex. | | Descriptor: | (3S)-TETRAHYDROFURAN-3-YL (1R)-3-{(2R)-4-[(1S,3S)-3-(2-AMINO-2-OXOETHYL)-2,3-DIHYDRO-1H-INDEN-1-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1V7Y
 
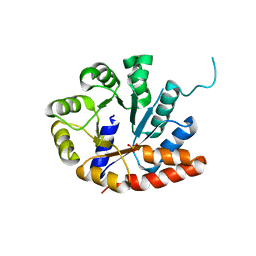 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli at room temperature | | Descriptor: | SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-25 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
1URA
 
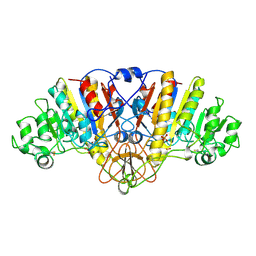 | | ALKALINE PHOSPHATASE (D51ZN) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Tibbitts, T.T, Murphy, J.E, Kantrowitz, E.R. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Kinetic and structural consequences of replacing the aspartate bridge by asparagine in the catalytic metal triad of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 257, 1996
|
|
2B8V
 
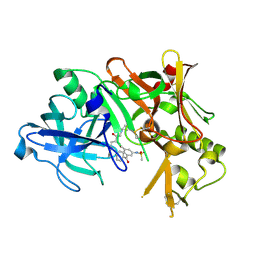 | | Crystal structure of human Beta-secretase complexed with L-L000430,469 | | Descriptor: | 3-BENZOYL-N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]BENZAMIDE, Beta-secretase 1 | | Authors: | Stachel, S.J, Coburn, C.A, Steele, T.G, Crouthamel, M.-C, Pietrak, B.L, Lai, M.-T, Holloway, M.K, Munshi, S.K, Graham, S.L, Vacca, J.P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally biased P3 amide replacements of beta-secretase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2B99
 
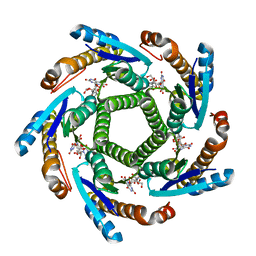 | | Crystal Structure of an archaeal pentameric riboflavin synthase Complex with a Substrate analog inhibitor | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
