2X6O
 
 | | Tet Repressor class D in complex with 7-chlor-2-cyano-iso- tetracycline | | Descriptor: | (4S,4AS,6S,8AS)-6-[(1S)-7-CHLORO-4-HYDROXY-1-METHYL-3-OXO-1,3-DIHYDRO-2-BENZOFURAN-1-YL]-4-(DIMETHYLAMINO)-3,8A-DIHYDROXY-1,8-DIOXO-1,4,4A,5,6,7,8,8A-OCTAHYDRONAPHTHALENE-2-CARBONITRILE, CHLORIDE ION, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Volkers, G, Hinrichs, W. | | Deposit date: | 2010-02-18 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of Drug Degradation Products by Target Proteins: Isotetracycline Binding to Tet Repressor.
J.Med.Chem., 54, 2011
|
|
8B4A
 
 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
8FTO
 
 | |
2VKE
 
 | |
2X9D
 
 | |
3Q3S
 
 | |
1JFS
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
7P6X
 
 | | Cryo-Em structure of the hexameric RUVBL1-RUVBL2 in complex with ZNHIT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Llorca, O, Serna, M, Fernandez-Leiro, R. | | Deposit date: | 2021-07-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of RUVBL1-RUVBL2-ZNHIT2, a complex that interacts with pre-mRNA-processing-splicing factor 8.
Nucleic Acids Res., 50, 2022
|
|
9EW2
 
 | | High resolution structure of FZD7 in complex with miniGs protein | | Descriptor: | Frizzled-7, GNAS complex locus,Isoform 4 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Ggamma, ... | | Authors: | Bous, J, Kinsolving, J, Gratz, L, Scharf, M.M, Voss, J, Selcuk, B, Adebali, O, Schulte, G. | | Deposit date: | 2024-04-03 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High resolution structure of FZD7 in complex with miniGs protein
To Be Published
|
|
9EPO
 
 | | High resolution structure of FZD7 in inactive conformation | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Frizzled-7, PALMITIC ACID | | Authors: | Bous, J, Kinsolving, J, Gratz, L, Scharf, M.M, Voss, J, Selcuk, B, Adebali, O, Schulte, G. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | high-resolution structural characterization of FZD7 unravels the importance of water network and cholesterol for class F GPCRs dynamic and activation
To Be Published
|
|
7ABZ
 
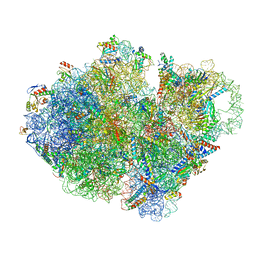 | | Structure of pre-accomodated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
1DDL
 
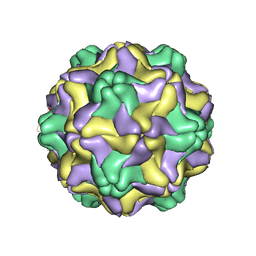 | | DESMODIUM YELLOW MOTTLE TYMOVIRUS | | Descriptor: | DESMODIUM YELLOW MOTTLE VIRUS, RNA (5'-R(P*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Larson, S.B, Day, J, Canady, M.A, Greenwood, A, McPherson, A. | | Deposit date: | 1999-11-10 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Refined structure of desmodium yellow mottle tymovirus at 2.7 A resolution.
J.Mol.Biol., 301, 2000
|
|
5UYN
 
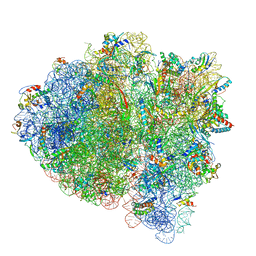 | | 70S ribosome bound with near-cognate ternary complex not base-paired to A site codon (Structure I-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5UYM
 
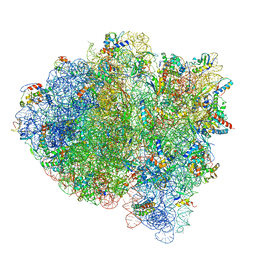 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon, closed 30S (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5UYQ
 
 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, closed 30S (Structure III-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5UYL
 
 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity.
Nature, 546, 2017
|
|
5UYP
 
 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, open 30S (Structure II-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
8HMX
 
 | |
8I9T
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9R
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State 5S RNP | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9X
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8IA0
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Puf6 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9W
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Dbp10-3 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9Y
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
4YBB
 
 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
