6Q5J
 
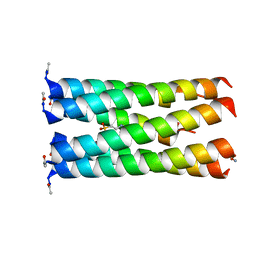 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24E | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24E | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5M
 
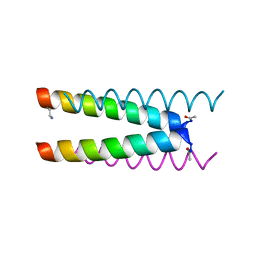 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24Dab | | Descriptor: | CC-Hex*-L24Dab, SODIUM ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
4X7V
 
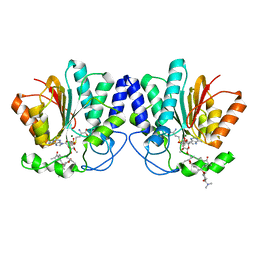 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin IV (product) | | Descriptor: | MAGNESIUM ION, MYCINAMICIN IV, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
5EBJ
 
 | | Joint X-ray/neutron structure of reversibly photoswitching chromogenic protein, Dathail | | Descriptor: | photoswitching chromogenic protein | | Authors: | Kovalevsky, A.Y, Langan, P.S, Bradbury, A.R.M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail.
J.Mol.Biol., 428, 2016
|
|
6KM3
 
 | | Human Carbonic Anhydrase II native 00 atm CO2 | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6QLT
 
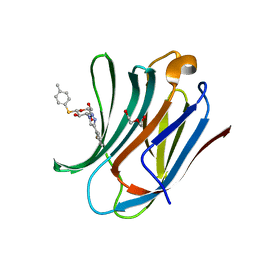 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-7 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, DI(HYDROXYETHYL)ETHER, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
7FCI
 
 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
5EJU
 
 | |
6Q5S
 
 | | Crystal structure of a de novo designed antiparallel four-helix coiled coil apCC-Tet | | Descriptor: | apCC-Tet | | Authors: | Beesley, J.L, Guto, G.R, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
3G30
 
 | |
4V8L
 
 | |
4V65
 
 | | Structure of the E. coli ribosome in the Pre-accommodation state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Devkota, B, Caulfield, T.R, Tan, R.-Z, Harvey, S.C. | | Deposit date: | 2008-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Structure of the E. coli Ribosome Before and After Accommodation: Implications for Proofreading
To be Published
|
|
5VUF
 
 | | HLA-B*57:01 presenting LTVQVARVY | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2017-05-19 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome.
Nat Commun, 9, 2018
|
|
5VWJ
 
 | | HLA-B*58:01 presenting LTVQVARVW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-58 alpha chain, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2017-05-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome.
Nat Commun, 9, 2018
|
|
4XJB
 
 | | X-ray structure of Lysozyme1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJG
 
 | | X-ray structure of Lysozyme B2 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJI
 
 | | X-ray structure of LysozymeS2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6QLO
 
 | | Galectin-3C in complex with substituted fluoroaryltriazole monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[3,4-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6QLQ
 
 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative 4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
3G31
 
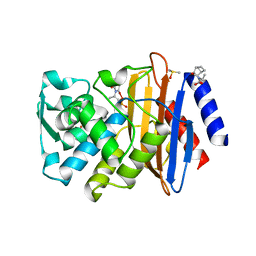 | | CTX-M-9 class A beta-lactamase complexed with compound 4 (GF1) | | Descriptor: | (2S)-2-[(3aR,4R,7S,7aS)-1,3-dioxooctahydro-2H-4,7-methanoisoindol-2-yl]propanoic acid, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
5W2S
 
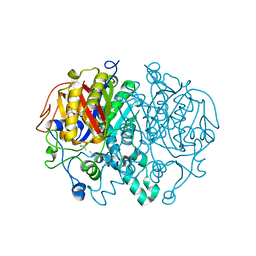 | |
4XJD
 
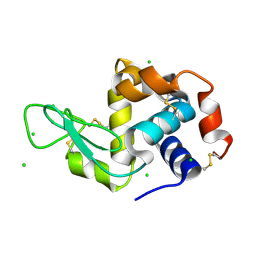 | | X-ray structure of Lysozyme2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJH
 
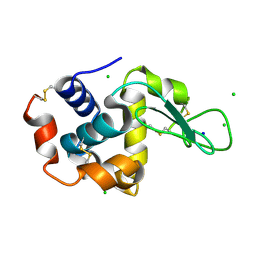 | | X-ray structure of LysozymeS1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6Q5I
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24E | | Descriptor: | AMMONIUM ION, CC-Hex*-L24E, SULFATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
4XOK
 
 | | Observing the overall rocking motion of a protein in a crystal. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Ma, P, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
