1SF7
 
 | | BINDING OF TETRA-N-ACETYLCHITOTETRAOSE TO HEW LYSOZYME: A POWDER DIFFRACTION STUDY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Von Dreele, R.B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-02 | | Last modified: | 2020-07-29 | | Method: | POWDER DIFFRACTION | | Cite: | Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: a powder diffraction study.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4KRT
 
 | |
1ENJ
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
3MUZ
 
 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M.L, Driedger, M.R, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
4XHB
 
 | |
4KTF
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4,4'-(3-amino-1H-pyrazole-4,5-diyl)diphenol | | Descriptor: | 4,4'-(3-amino-1H-pyrazole-4,5-diyl)diphenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2013-05-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Overcoming the Limitations of Fragment Merging: Rescuing a Strained Merged Fragment Series Targeting Mycobacterium tuberculosis CYP121.
Chemmedchem, 8, 2013
|
|
1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-10-24 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
4XJ2
 
 | | FerA - NADH:FMN oxidoreductase from Paracoccus denitrificans in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase domain protein, FMN-binding protein | | Authors: | Sedlacek, V, Klumpler, T, Marek, J, Kucera, I. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical properties and crystal structure of the flavin reductase FerA from Paracoccus denitrificans.
Microbiol. Res., 188-189, 2016
|
|
1SJ2
 
 | | Crystal structure of Mycobacterium tuberculosis catalase-peroxidase | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase/catalase T | | Authors: | Bertrand, T, Eady, N.A.J, Jones, J.N, Bodiguel, J, Jesmin, Nagy, J.M, Raven, E.L, Jamart-Gregoire, B, Brown, K.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Catalase-Peroxidase.
J.Biol.Chem., 279, 2004
|
|
4KET
 
 | | Crystal structure of SsoPox W263I | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
5HSA
 
 | | Alcohol Oxidase AOX1 from Pichia Pastoris | | Descriptor: | ARABINO-FLAVIN-ADENINE DINUCLEOTIDE, Alcohol oxidase 1, CALCIUM ION, ... | | Authors: | Neumann, P, Ficner, R, Feussner, I, Koch, C. | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alcohol Oxidase from Pichia pastoris.
Plos One, 11, 2016
|
|
1E7M
 
 | |
4XAR
 
 | | mGluR2 ECD and mGluR3 ECD complex with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, IODIDE ION, Metabotropic glutamate receptor 3 | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
4KPA
 
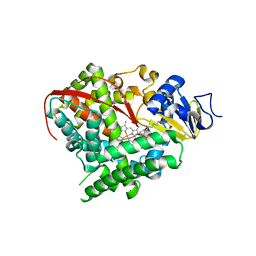 | | Crystal structure of cytochrome P450 BM-3 in complex with N-palmitoylglycine (NPG) | | Descriptor: | Cytochrome P450 BM-3, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Amadeo, G.A, Catalano, J, McDermott, A.E, Tong, L. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence: A Single Charged Residue Affects Substrate Binding in Cytochrome P450 BM-3.
Biochemistry, 52, 2013
|
|
4KPR
 
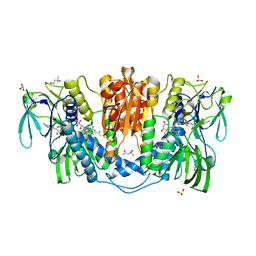 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
1SLI
 
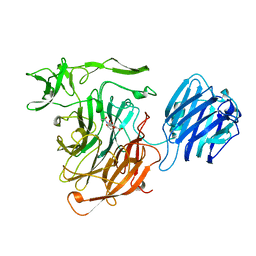 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Chou, M.Y, Li, Y.T, Luo, M. | | Deposit date: | 1998-03-28 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity.
Structure, 6, 1998
|
|
4XBR
 
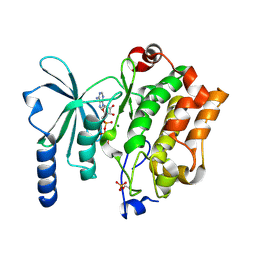 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
5HY1
 
 | | high resolution structure of barbiturase | | Descriptor: | 1,3,5-triazine-2,4,6-triol, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Scott, C, Balotra, S, Wilding, M, Newman, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
1EA0
 
 | | Alpha subunit of A. brasilense glutamate synthase | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Binda, C, Bossi, R.T, Vanoni, M.A, Mattevi, A. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-Talk and Ammonia Channeling between Active Centers in the Unexpected Domain Arrangement of Glutamate Synthase
Structure, 8, 2000
|
|
5HXU
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
4KTZ
 
 | | Lactococcus phage 67 RuvC | | Descriptor: | MAGNESIUM ION, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
4L1Q
 
 | |
4XAD
 
 | |
1F14
 
 | |
3MU7
 
 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
