6QA3
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, PYRAZOLE, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
1S0Y
 
 | | The structure of trans-3-chloroacrylic acid dehalogenase, covalently inactivated by the mechanism-based inhibitor 3-bromopropiolate at 2.3 Angstrom resolution | | Descriptor: | MALONIC ACID, alpha-subunit of trans-3-chloroacrylic acid dehalogenase, beta-subunit of trans-3-chloroacrylic acid dehalogenase | | Authors: | de Jong, R.M, Brugman, W, Poelarends, G.J, Whitman, C.P, Dijkstra, B.W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of trans-3-chloroacrylic acid dehalogenase reveals a novel hydration mechanism in the tautomerase superfamily
J.Biol.Chem., 279, 2004
|
|
4V8B
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-06 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4W2F
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with amicoumacin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4V9N
 
 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
1S38
 
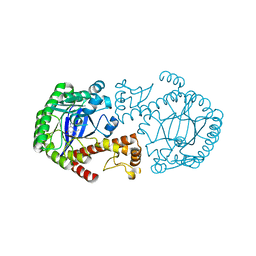 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Meyer, E.A, Furler, M, Diederich, F, Brenk, R, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis and In vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
1S3M
 
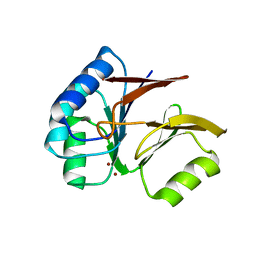 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, NICKEL (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
6CSW
 
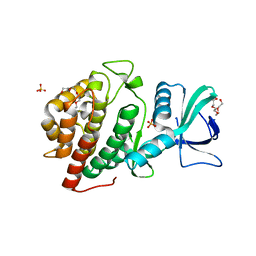 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor | | Descriptor: | (7R)-5-butyl-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7,8-dimethyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Chiodi, C.G, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor
To Be Published
|
|
6QA1
 
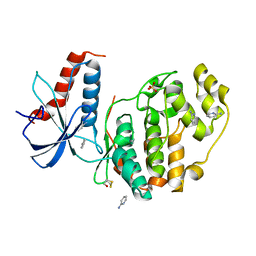 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, pyridin-2-amine | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
1SFF
 
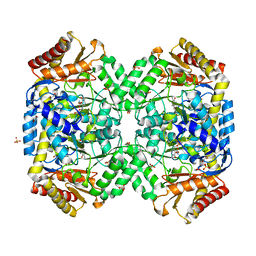 | | Structure of gamma-aminobutyrate aminotransferase complex with aminooxyacetate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
6QAG
 
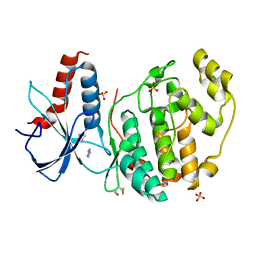 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-1,2,3-triazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6QA4
 
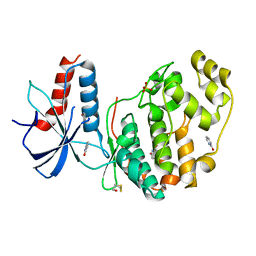 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-pyridin-2-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6CMM
 
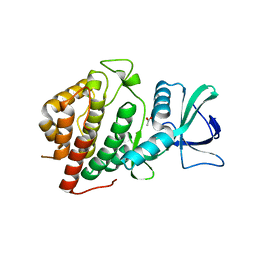 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor
To Be Published
|
|
1S3N
 
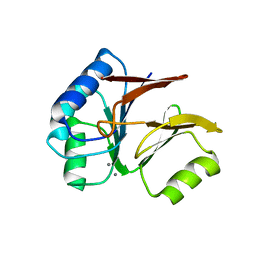 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, MANGANESE (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
6CPW
 
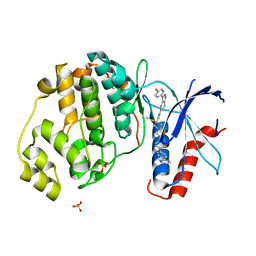 | | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology | | Descriptor: | (3S)-N-[3-(4-fluorophenyl)-1H-indazol-5-yl]-3-(methylsulfanyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Hruza, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4GBT
 
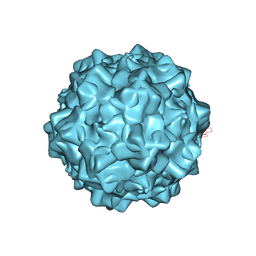 | | Structural characterization of H-1 Parvovirus: comparison of infectious virions to replication defective particles | | Descriptor: | CHLORIDE ION, Capsid protein VP1, SODIUM ION | | Authors: | Halder, S, Nam, H.-J, Govindasamy, L, Vogel, M, Dinsart, C, Salome, N, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2012-07-27 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural characterization of h-1 parvovirus: comparison of infectious virions to empty capsids.
J.Virol., 87, 2013
|
|
7P3K
 
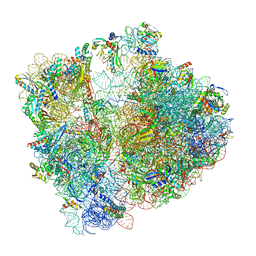 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide (control) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Buschauer, R, Komar, T, Becker, T, Berninghausen, O, Cheng, J, Beckmann, R. | | Deposit date: | 2021-07-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
4V6A
 
 | | Structure of EF-P bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome.
Science, 325, 2009
|
|
6Z6M
 
 | | Cryo-EM structure of human 80S ribosomes bound to EBP1, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
7SNZ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((2-((4-cyanophenyl)amino)pyrimidin-4-yl)amino)-5,7-dimethylindolizine-2-carbonitrile (JLJ604) | | Descriptor: | (4R)-6-{[2-(4-cyanoanilino)pyrimidin-4-yl]amino}-5,7-dimethylindolizine-2-carbonitrile, 1,4-DIAMINOBUTANE, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
4FSJ
 
 | | Crystal structure of the virus like particle of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
7SO6
 
 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SNP
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-(2-morpholinoethoxy)phenoxy)phenyl)acrylonitrile (JLJ530) | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-[2-(morpholin-4-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO2
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase K103N/Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.767 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
