1E9S
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Unbound monoclinic form. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
3C98
 
 | | Revised structure of the munc18a-syntaxin1 complex | | Descriptor: | Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Hattendorf, D.A, Misura, K.M, Burkhardt, P, Scheller, R.H, Fasshauer, D, Weis, W.I. | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide
Embo J., 27, 2008
|
|
5OY1
 
 | |
5OY2
 
 | | Direct-evolutioned unspecific peroxygenase from Agrocybe aegerita, in complex with DMP | | Descriptor: | 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2017-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Insights into the Substrate Promiscuity of a Laboratory-Evolved Peroxygenase.
Acs Chem.Biol., 13, 2018
|
|
3UAL
 
 | |
8PNM
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 2 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
8PNR
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 1 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
3UBW
 
 | | Complex of 14-3-3 isoform epsilon, a Mlf1 phosphopeptide and a small fragment hit from a FBDD screen | | Descriptor: | (3S)-pyrrolidin-3-ol, 14-3-3 protein epsilon, Myeloid leukemia factor 1, ... | | Authors: | Molzan, M, Weyand, M, Rose, R, Ottmann, C. | | Deposit date: | 2011-10-25 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the MLF1/14-3-3 interaction.
Febs J., 279, 2012
|
|
2LCK
 
 | |
2EFZ
 
 | |
5XY4
 
 | | CATPO mutant - V536W | | Descriptor: | CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase, ... | | Authors: | Yuzugullu Karakus, Y, Goc, G, Balci, S, Pearson, A.R, Yorke, B. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of the site of oxidase substrate binding in Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6AD3
 
 | |
6ASX
 
 | | CryoEM structure of E.coli his pause elongation complex | | Descriptor: | DNA (32-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Landick, R, Darst, S.A. | | Deposit date: | 2017-08-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing.
Mol. Cell, 69, 2018
|
|
5Y17
 
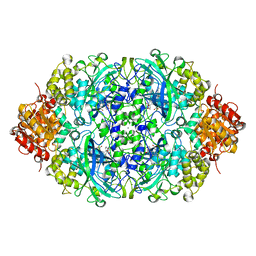 | | CATPO mutant - E316F | | Descriptor: | CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase | | Authors: | Karakus Yuzugullu, Y, Goc, G, Balci, S, Pearson, A.R, Yorke, B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the site of oxidase substrate binding in Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7VJT
 
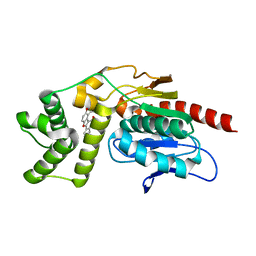 | | Crystal Structure of Mtb Pks13-TE in complex with inhibitor coumestan derivative 8 | | Descriptor: | 3,8-bis(oxidanyl)-7-(piperidin-1-ylmethyl)-[1]benzofuro[3,2-c]chromen-6-one, Polyketide synthase Pks13 (Termination polyketide synthase) | | Authors: | Zhang, W, Wang, S.S, Yu, L.F. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of Coumestan Derivatives as Polyketide Synthase 13-Thioesterase(Pks13-TE) Inhibitors with Improved hERG Profiles for Mycobacterium tuberculosis Treatment.
J.Med.Chem., 65, 2022
|
|
3SYY
 
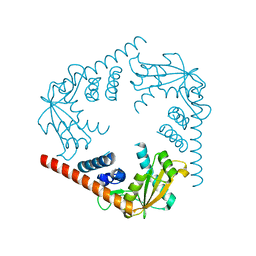 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
3SZ4
 
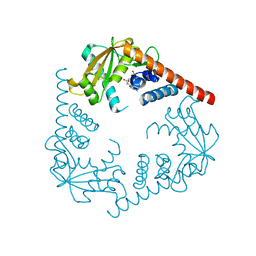 | | Crystal Structure of LHK-Exo in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3TDA
 
 | | Competitive replacement of thioridazine by prinomastat in crystals of cytochrome P450 2D6 | | Descriptor: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, Prinomastat, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
2LV6
 
 | | The complex between Ca-Calmodulin and skeletal muscle myosin light chain kinase from combination of NMR and aqueous and contrast-matched SAXS data | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase 2, ... | | Authors: | Grishaev, A.V, Anthis, N.J, Clore, G.M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Contrast-matched small-angle X-ray scattering from a heavy-atom-labeled protein in structure determination: application to a lead-substituted calmodulin-peptide complex.
J.Am.Chem.Soc., 134, 2012
|
|
7X2U
 
 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | Authors: | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-02-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
3SZ5
 
 | | Crystal Structure of LHK-Exo in complex with 5-phosphorylated oligothymidine (dT)4 | | Descriptor: | 5'-D(P*TP*TP*TP*T)-3', Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
2K0E
 
 | | A Coupled Equilibrium Shift Mechanism in Calmodulin-Mediated Signal Transduction | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Gsponer, J, Christodoulou, J, Cavalli, A, Bui, J.M, Richter, B, Dobson, C.M, Vendruscolo, M. | | Deposit date: | 2008-02-02 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction
Structure, 16, 2008
|
|
2JZI
 
 | |
2K0F
 
 | | Calmodulin complexed with calmodulin-binding peptide from smooth muscle myosin light chain kinase | | Descriptor: | 19-mer peptide from Myosin light chain kinase, CALCIUM ION, calmodulin | | Authors: | Gsponer, J, Christodoulou, J, Cavalli, A, Bui, J.M, Richter, B, Dobson, C.M, Vendruscolo, M. | | Deposit date: | 2008-02-02 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction
Structure, 16, 2008
|
|
