8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8K7E
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3R,4S,5R)-2-(aminomethyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(aminomethyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
8K7L
 
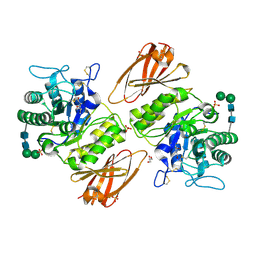 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3R,4S,5R)-2-(aminomethyl)-5-(hydroxymethyl)-1-methylpyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(aminomethyl)-5-(hydroxymethyl)-1-methyl-pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
6CM4
 
 | | Structure of the D2 Dopamine Receptor Bound to the Atypical Antipsychotic Drug Risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, D(2) dopamine receptor, endolysin chimera, ... | | Authors: | Wang, S, Che, T, Levit, A, Shoichet, B.K, Wacker, D, Roth, B.L. | | Deposit date: | 2018-03-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone.
Nature, 555, 2018
|
|
8CE1
 
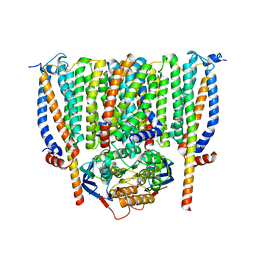 | | Cytochrome c maturation complex CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
6VBY
 
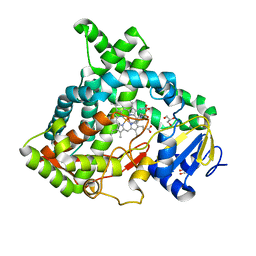 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
8CEA
 
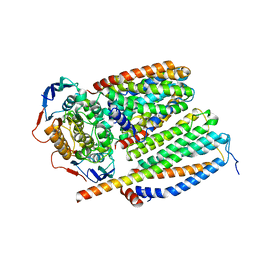 | | Cytochrome c maturation complex CcmABCD, E154Q | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
3B9L
 
 | | Human serum albumin complexed with myristate and AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, MYRISTIC ACID, Serum albumin | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
6IQU
 
 | |
6VCN
 
 | | Crystal structure of E.coli RppH in complex with ppcpG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
8OF1
 
 | | Structure of ALDH5F1 from moss Physcomitrium patens in complex with NAD+ in the contracted conformation | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
8BRT
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
8BRJ
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
6V6Y
 
 | | Crystal Structure of T. thermophilus methylenetetrahydrofolate dehydrogenase (MTHFD) | | Descriptor: | Bifunctional protein FolD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maiello, F, Coelho, C, Gallo, G, Sucharski, F, Hardy, L, Wurtele, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15201569 Å) | | Cite: | Crystal structure of Thermus thermophilus methylenetetrahydrofolate dehydrogenase and determinants of thermostability.
Plos One, 15, 2020
|
|
8BNU
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
6V7N
 
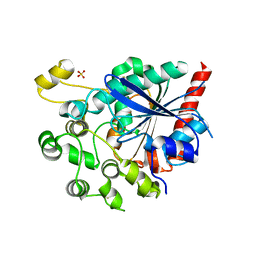 | |
6M7X
 
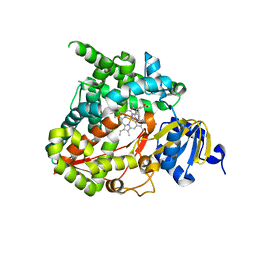 | | Structure of human CYP11B1 in complex with fadrozole | | Descriptor: | 4-[(5S)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Structure of human cortisol-producing cytochrome P450 11B1 bound to the breast cancer drug fadrozole provides insights for drug design.
J. Biol. Chem., 294, 2019
|
|
4ZLE
 
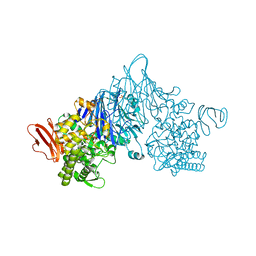 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
8K7J
 
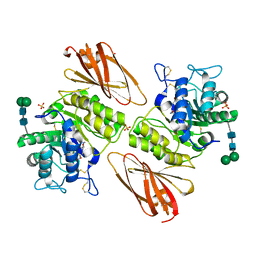 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3R,4S,5R)-2-((dimethylamino)methyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[(dimethylamino)methyl]-5-(hydroxymethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
8K7F
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3S,4R,5R)-2,5-bis(hydroxymethyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2,5-bis(hydroxymethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
6COM
 
 | | 2.3A crystal structure of E. coli phosphoenolpyruvate carboxykinase mutant Asp269Asn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Sokaribo, A.S, Cotelesage, J.H, Novakovski, B, Goldie, H, Sanders, D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
8BRS
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
8K7G
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3S,4R,5R)-2,5-bis(hydroxymethyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2,5-bis(hydroxymethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
