4UYO
 
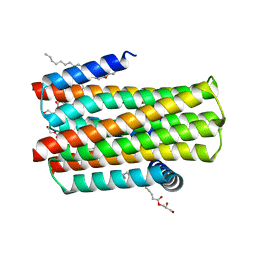 | | Structure of delta7-DgkA in 7.9 MAG by serial femtosecond crystatallography to 2.18 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CITRATE ANION, ... | | Authors: | Li, D, Howe, N, Other, O, Caffrey, M. | | Deposit date: | 2014-09-02 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
3SFC
 
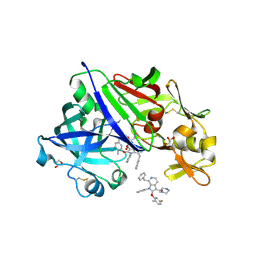 | | Structure-Based Optimization of Potent 4- and 6-Azaindole-3-Carboxamides as Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Renin, ... | | Authors: | Scheiper, B, Matter, H, Steinhagen, H, Bocskei, Z, Fleury, V, McCort, G. | | Deposit date: | 2011-06-13 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of potent 4- and 6-azaindole-3-carboxamides as renin inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4UXX
 
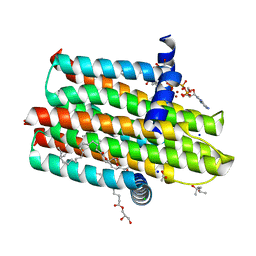 | | Structure of delta4-DgkA with AMPPCP in 9.9 MAG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Li, D, Vogeley, L, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
3S9N
 
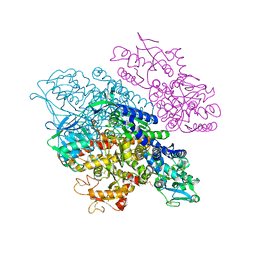 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V2W
 
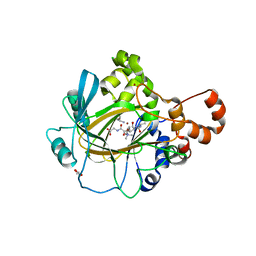 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (16-35) | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, Zafred, D, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
4V1U
 
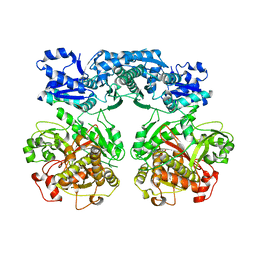 | |
4W5S
 
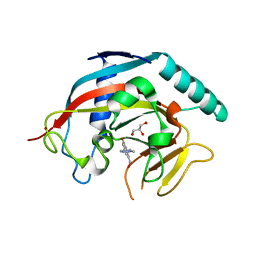 | | Tankyrase in complex with compound | | Descriptor: | 8-(hydroxymethyl)-2-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]quinazolin-4(3H)-one, GLYCEROL, Tankyrase-1, ... | | Authors: | Johannes, J, Kazmirski, S.L, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
4W7M
 
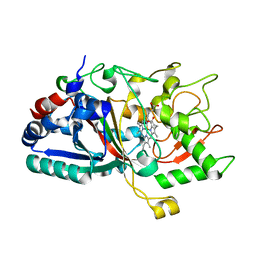 | |
4UVA
 
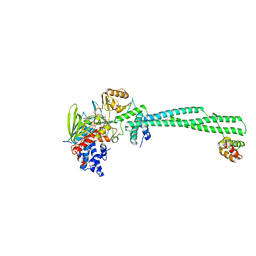 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1R,2S) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS)-4a-[(1S,3E)-3-imino-1-phenylbutyl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
7MZX
 
 | |
4V0K
 
 | |
4V1T
 
 | |
3S8R
 
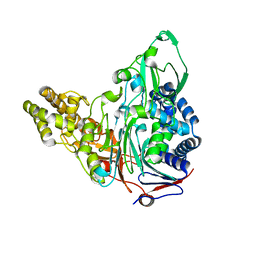 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | GLYCEROL, Glutaryl-7-aminocephalosporanic-acid acylase | | Authors: | Kim, J.K, Yang, I.S, Park, S.S, Kim, K.H. | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation.
Biochemistry, 42, 2003
|
|
4V1V
 
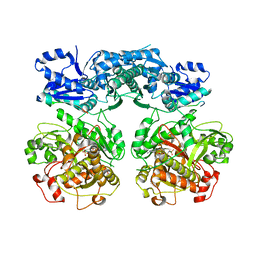 | | Heterocyclase in complex with substrate and Cofactor | | Descriptor: | LYND, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Koehnke, J, Naismith, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Analysis of Leader Peptide Binding Enables Leader-Free Cyanobactin Processing.
Nat.Chem.Biol., 11, 2015
|
|
3J2P
 
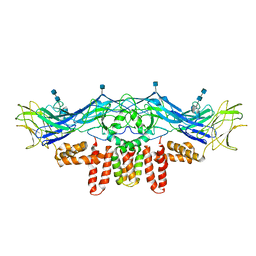 | | CryoEM structure of Dengue virus envelope protein heterotetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
4V0O
 
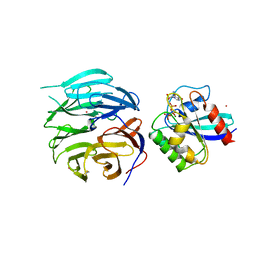 | |
4UZ7
 
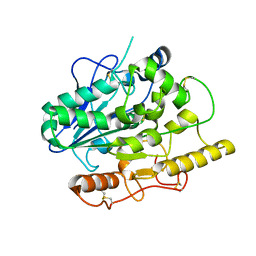 | |
4V0L
 
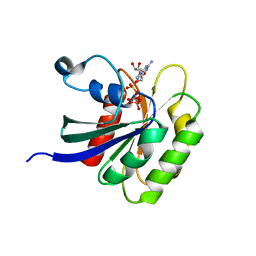 | |
3S97
 
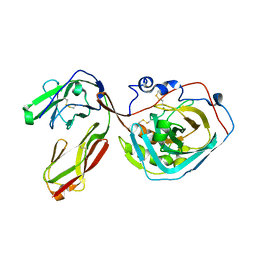 | | PTPRZ CNTN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Bouyain, S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2971 Å) | | Cite: | A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4WF0
 
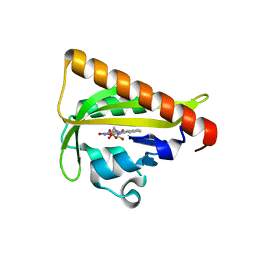 | |
4WGZ
 
 | |
3S29
 
 | |
3S4B
 
 | | Cellobiose phosphorylase from Cellulomonas uda in complex with glucose | | Descriptor: | Cellobiose phosphorylase, alpha-D-glucopyranose | | Authors: | Van Hoorebeke, A, Stout, J, Soetaert, W, Van Beeumen, J, Desmet, T, Savvides, S. | | Deposit date: | 2011-05-19 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellobiose phosphorylase: reconstructing the structural itinerary along the catalytic pathway
To be Published
|
|
3SEK
 
 | | Crystal Structure of the Myostatin:Follistatin-like 3 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Follistatin-related protein 3, Growth/differentiation factor 8 | | Authors: | Cash, J.N, Thompson, T.B. | | Deposit date: | 2011-06-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of myostatinfollistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding.
J.Biol.Chem., 287, 2012
|
|
3S9L
 
 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, cryocooled 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
